
Nesterenkonia jeotgali
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Nesterenkonia
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
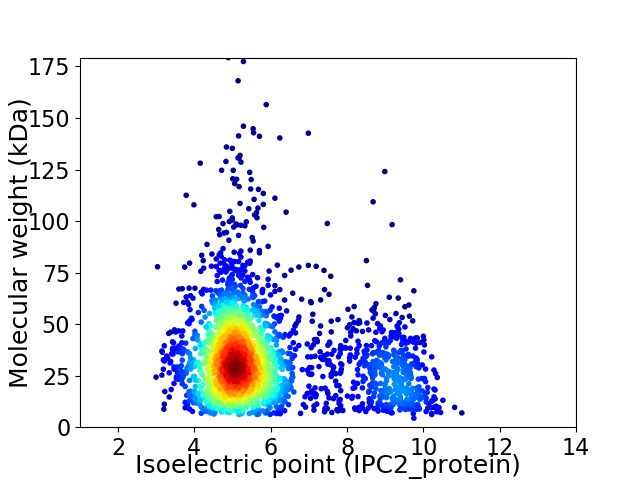
Virtual 2D-PAGE plot for 2504 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0W8ICD3|A0A0W8ICD3_9MICC Acyl-CoA_dh_2 domain-containing protein OS=Nesterenkonia jeotgali OX=317018 GN=AVL63_13120 PE=4 SV=1
MM1 pKa = 7.47TGPSPRR7 pKa = 11.84HH8 pKa = 5.43LWSSSLALTMLLLVTSSCSAEE29 pKa = 3.73QSAQNPSEE37 pKa = 4.23QEE39 pKa = 3.62PSAQDD44 pKa = 3.44SDD46 pKa = 3.91SSPPAEE52 pKa = 4.0QRR54 pKa = 11.84SSPSSAAVGAEE65 pKa = 3.93DD66 pKa = 4.21PEE68 pKa = 4.43EE69 pKa = 3.86EE70 pKa = 4.32AEE72 pKa = 4.24AVTEE76 pKa = 4.2VEE78 pKa = 4.24EE79 pKa = 4.32EE80 pKa = 4.13EE81 pKa = 4.05PSIRR85 pKa = 11.84DD86 pKa = 3.26VEE88 pKa = 4.29FADD91 pKa = 5.09LEE93 pKa = 4.15WTEE96 pKa = 4.03WVNEE100 pKa = 4.15VVLIPAEE107 pKa = 4.45GSDD110 pKa = 3.7GPEE113 pKa = 3.72FEE115 pKa = 4.54VSEE118 pKa = 4.61EE119 pKa = 3.55IHH121 pKa = 6.33YY122 pKa = 11.24ADD124 pKa = 4.93ADD126 pKa = 4.07GDD128 pKa = 4.0GHH130 pKa = 6.77EE131 pKa = 5.17DD132 pKa = 3.49ALVSLGWTDD141 pKa = 3.85DD142 pKa = 4.04RR143 pKa = 11.84GFEE146 pKa = 4.6DD147 pKa = 4.51IWYY150 pKa = 8.61IWAWDD155 pKa = 3.89PDD157 pKa = 4.13TEE159 pKa = 4.46SAYY162 pKa = 10.32QAGGPVARR170 pKa = 11.84AANCGDD176 pKa = 3.35HH177 pKa = 6.5VEE179 pKa = 4.39SVSAGEE185 pKa = 3.74DD186 pKa = 3.06HH187 pKa = 7.27FVIEE191 pKa = 4.09EE192 pKa = 4.16HH193 pKa = 6.58LWPQNADD200 pKa = 3.34HH201 pKa = 7.63DD202 pKa = 4.62CASTPSIQVTRR213 pKa = 11.84HH214 pKa = 5.65LVLEE218 pKa = 4.67DD219 pKa = 3.91DD220 pKa = 4.31WPVMISPVRR229 pKa = 11.84AHH231 pKa = 7.11GGMCPQILGTDD242 pKa = 3.86DD243 pKa = 3.7LWPVEE248 pKa = 4.2DD249 pKa = 4.7FEE251 pKa = 5.65FRR253 pKa = 11.84TSPEE257 pKa = 3.55EE258 pKa = 3.72AAGLVGDD265 pKa = 4.64PAPTAFAEE273 pKa = 4.13ADD275 pKa = 3.23VWSRR279 pKa = 11.84LWLHH283 pKa = 5.89YY284 pKa = 9.35PNWMLVHH291 pKa = 6.73LAYY294 pKa = 10.09MNPADD299 pKa = 4.62GPPVLEE305 pKa = 5.31GGYY308 pKa = 7.65TPCGWVYY315 pKa = 10.96LEE317 pKa = 4.49EE318 pKa = 4.28DD319 pKa = 3.97TRR321 pKa = 11.84PVPHH325 pKa = 7.35DD326 pKa = 4.23PYY328 pKa = 10.68PPEE331 pKa = 4.28DD332 pKa = 3.25
MM1 pKa = 7.47TGPSPRR7 pKa = 11.84HH8 pKa = 5.43LWSSSLALTMLLLVTSSCSAEE29 pKa = 3.73QSAQNPSEE37 pKa = 4.23QEE39 pKa = 3.62PSAQDD44 pKa = 3.44SDD46 pKa = 3.91SSPPAEE52 pKa = 4.0QRR54 pKa = 11.84SSPSSAAVGAEE65 pKa = 3.93DD66 pKa = 4.21PEE68 pKa = 4.43EE69 pKa = 3.86EE70 pKa = 4.32AEE72 pKa = 4.24AVTEE76 pKa = 4.2VEE78 pKa = 4.24EE79 pKa = 4.32EE80 pKa = 4.13EE81 pKa = 4.05PSIRR85 pKa = 11.84DD86 pKa = 3.26VEE88 pKa = 4.29FADD91 pKa = 5.09LEE93 pKa = 4.15WTEE96 pKa = 4.03WVNEE100 pKa = 4.15VVLIPAEE107 pKa = 4.45GSDD110 pKa = 3.7GPEE113 pKa = 3.72FEE115 pKa = 4.54VSEE118 pKa = 4.61EE119 pKa = 3.55IHH121 pKa = 6.33YY122 pKa = 11.24ADD124 pKa = 4.93ADD126 pKa = 4.07GDD128 pKa = 4.0GHH130 pKa = 6.77EE131 pKa = 5.17DD132 pKa = 3.49ALVSLGWTDD141 pKa = 3.85DD142 pKa = 4.04RR143 pKa = 11.84GFEE146 pKa = 4.6DD147 pKa = 4.51IWYY150 pKa = 8.61IWAWDD155 pKa = 3.89PDD157 pKa = 4.13TEE159 pKa = 4.46SAYY162 pKa = 10.32QAGGPVARR170 pKa = 11.84AANCGDD176 pKa = 3.35HH177 pKa = 6.5VEE179 pKa = 4.39SVSAGEE185 pKa = 3.74DD186 pKa = 3.06HH187 pKa = 7.27FVIEE191 pKa = 4.09EE192 pKa = 4.16HH193 pKa = 6.58LWPQNADD200 pKa = 3.34HH201 pKa = 7.63DD202 pKa = 4.62CASTPSIQVTRR213 pKa = 11.84HH214 pKa = 5.65LVLEE218 pKa = 4.67DD219 pKa = 3.91DD220 pKa = 4.31WPVMISPVRR229 pKa = 11.84AHH231 pKa = 7.11GGMCPQILGTDD242 pKa = 3.86DD243 pKa = 3.7LWPVEE248 pKa = 4.2DD249 pKa = 4.7FEE251 pKa = 5.65FRR253 pKa = 11.84TSPEE257 pKa = 3.55EE258 pKa = 3.72AAGLVGDD265 pKa = 4.64PAPTAFAEE273 pKa = 4.13ADD275 pKa = 3.23VWSRR279 pKa = 11.84LWLHH283 pKa = 5.89YY284 pKa = 9.35PNWMLVHH291 pKa = 6.73LAYY294 pKa = 10.09MNPADD299 pKa = 4.62GPPVLEE305 pKa = 5.31GGYY308 pKa = 7.65TPCGWVYY315 pKa = 10.96LEE317 pKa = 4.49EE318 pKa = 4.28DD319 pKa = 3.97TRR321 pKa = 11.84PVPHH325 pKa = 7.35DD326 pKa = 4.23PYY328 pKa = 10.68PPEE331 pKa = 4.28DD332 pKa = 3.25
Molecular weight: 36.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0W8II41|A0A0W8II41_9MICC Uncharacterized protein OS=Nesterenkonia jeotgali OX=317018 GN=AVL63_10775 PE=4 SV=1
MM1 pKa = 7.48RR2 pKa = 11.84AALFVLVFAVTVIGASLAGSQNLAFVLLMGAAVAALASTVMTASGRR48 pKa = 11.84QALRR52 pKa = 11.84RR53 pKa = 11.84TRR55 pKa = 11.84GNLTQGWSRR64 pKa = 11.84RR65 pKa = 11.84PGG67 pKa = 3.12
MM1 pKa = 7.48RR2 pKa = 11.84AALFVLVFAVTVIGASLAGSQNLAFVLLMGAAVAALASTVMTASGRR48 pKa = 11.84QALRR52 pKa = 11.84RR53 pKa = 11.84TRR55 pKa = 11.84GNLTQGWSRR64 pKa = 11.84RR65 pKa = 11.84PGG67 pKa = 3.12
Molecular weight: 6.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
820621 |
38 |
1656 |
327.7 |
35.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.617 ± 0.064 | 0.553 ± 0.012 |
5.623 ± 0.038 | 6.796 ± 0.054 |
3.036 ± 0.029 | 8.769 ± 0.043 |
2.267 ± 0.024 | 4.377 ± 0.031 |
2.204 ± 0.036 | 10.389 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.095 ± 0.022 | 2.067 ± 0.023 |
5.353 ± 0.035 | 3.649 ± 0.028 |
6.847 ± 0.044 | 6.15 ± 0.034 |
5.908 ± 0.029 | 7.935 ± 0.036 |
1.414 ± 0.019 | 1.95 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
