
Terrimicrobium sacchariphilum
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Terrimicrobia; Terrimicrobiales; Terrimicrobiaceae; Terrimicrobium
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
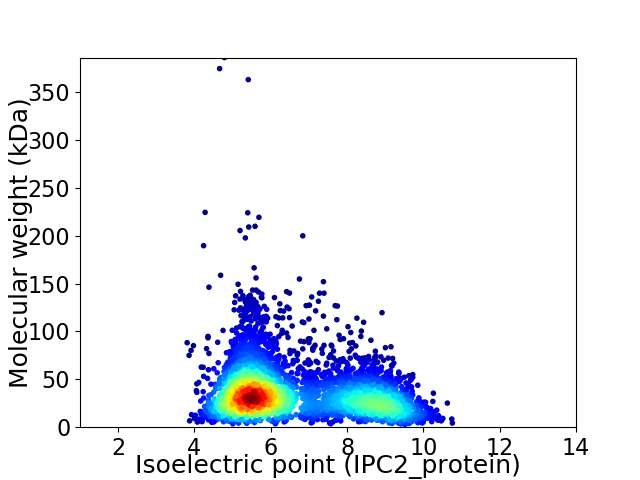
Virtual 2D-PAGE plot for 4168 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
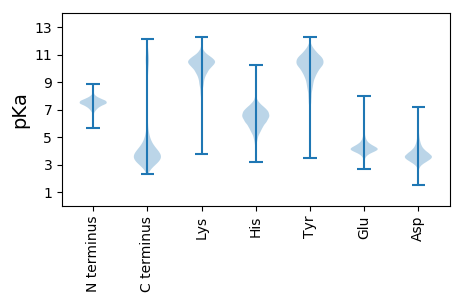
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A146GEE6|A0A146GEE6_TERSA UPF0234 protein TSACC_3114 OS=Terrimicrobium sacchariphilum OX=690879 GN=TSACC_3114 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 10.45GKK4 pKa = 10.05LARR7 pKa = 11.84TGGWIGVLSLLAAGMAAAAWNDD29 pKa = 3.47PVTNTPAIIQLDD41 pKa = 4.02FGPYY45 pKa = 9.37GNKK48 pKa = 10.26LPGDD52 pKa = 3.77GQNYY56 pKa = 9.52CEE58 pKa = 4.25PTSAVMSLFWLGSNGFTQLTPSTYY82 pKa = 10.0PGQNDD87 pKa = 4.27PYY89 pKa = 9.62TYY91 pKa = 10.86NLEE94 pKa = 4.05RR95 pKa = 11.84LMGGLMQTSATQGTYY110 pKa = 10.53DD111 pKa = 3.11SSAIQGISDD120 pKa = 3.56YY121 pKa = 10.97LAAAGLSGSFSYY133 pKa = 10.15IYY135 pKa = 10.69NGGDD139 pKa = 3.27SASNLLNPSWLAAQLAPNAGANPGSITFIDD169 pKa = 5.97FSISWYY175 pKa = 9.04GQNSGSSTSFSAEE188 pKa = 3.89GGHH191 pKa = 6.15EE192 pKa = 4.09LALLAVVDD200 pKa = 4.15PTGNAQLILNNAYY213 pKa = 9.61PSTLEE218 pKa = 3.81NVPNVPEE225 pKa = 4.95SDD227 pKa = 3.68PQTLGITQVPAGWNLPGMALPSEE250 pKa = 5.63DD251 pKa = 3.41YY252 pKa = 10.06WQLQSSDD259 pKa = 3.35GTVGDD264 pKa = 4.08GSTTKK269 pKa = 10.74AIVDD273 pKa = 3.52SAYY276 pKa = 10.57AFSVDD281 pKa = 3.16TMANPTLNPSWTPADD296 pKa = 3.52WVISGSKK303 pKa = 9.15TINTNSATFTVQAAVTGAGGFNKK326 pKa = 9.71TGEE329 pKa = 4.3GALVLTAGDD338 pKa = 4.6DD339 pKa = 3.77LTGNNAVSGGALGSTNAGDD358 pKa = 3.94STPLGTGSFVLTNGGTIGLTPAGSGGPLAFSIASGSGSSVTFGAGGGALEE408 pKa = 4.63IDD410 pKa = 3.16AGGYY414 pKa = 10.37DD415 pKa = 3.86SLSVTMGDD423 pKa = 4.02GSATSSNLIRR433 pKa = 11.84QSAGTFVIIPAHH445 pKa = 6.61GISALGTTEE454 pKa = 3.75RR455 pKa = 11.84VLVAGGSSSRR465 pKa = 11.84PASVNGMVAPYY476 pKa = 10.12IVGQDD481 pKa = 3.22NDD483 pKa = 4.26AGASGYY489 pKa = 9.82FLNYY493 pKa = 9.83DD494 pKa = 3.95GSAGFQKK501 pKa = 10.61AAAVSSLSTAIDD513 pKa = 3.51VATSSTVYY521 pKa = 10.44AAEE524 pKa = 4.11TDD526 pKa = 3.6QVIATNEE533 pKa = 3.96TARR536 pKa = 11.84VYY538 pKa = 10.91ALQVNDD544 pKa = 4.28SEE546 pKa = 5.08VSGTNATLEE555 pKa = 4.23VGSRR559 pKa = 11.84QAGDD563 pKa = 3.35SAGVILNGGTISASSLDD580 pKa = 3.51FGAAEE585 pKa = 4.09GVIYY589 pKa = 10.56ASEE592 pKa = 4.2AGGAISSTITSSAGLTTFGPGTVALSGDD620 pKa = 3.85STSTLSGTVTVNSGTLSAQSAQGSATGSANVSVQANATLSVSGAGAVAGNVSVQNNGTLLLSGGTISGNVTVAEE694 pKa = 4.5VGQSSANPGGTLAGSGTLSGSTTARR719 pKa = 11.84GVITSGASGGNITFEE734 pKa = 4.32GSADD738 pKa = 3.69LNSSQFFWQLDD749 pKa = 3.81SLTDD753 pKa = 3.82DD754 pKa = 4.24ASQAGIEE761 pKa = 4.19WNLLTFLGDD770 pKa = 3.44SHH772 pKa = 8.24VGSEE776 pKa = 4.28DD777 pKa = 3.71APVTIYY783 pKa = 10.9LQFAAGVITPDD794 pKa = 3.19TADD797 pKa = 4.11PFWMTDD803 pKa = 3.63HH804 pKa = 6.72EE805 pKa = 4.53WKK807 pKa = 10.2IWDD810 pKa = 4.1FSQATSYY817 pKa = 10.93SWDD820 pKa = 3.49DD821 pKa = 3.69SGDD824 pKa = 3.52GNTSFNYY831 pKa = 10.43GNFDD835 pKa = 3.83VVEE838 pKa = 4.27KK839 pKa = 10.76AADD842 pKa = 3.28NSVYY846 pKa = 10.43LKK848 pKa = 10.55YY849 pKa = 10.84VAVPEE854 pKa = 4.24PAVIWLALAGCGAMIASRR872 pKa = 11.84RR873 pKa = 11.84IRR875 pKa = 11.84QRR877 pKa = 11.84RR878 pKa = 3.26
MM1 pKa = 7.44KK2 pKa = 10.45GKK4 pKa = 10.05LARR7 pKa = 11.84TGGWIGVLSLLAAGMAAAAWNDD29 pKa = 3.47PVTNTPAIIQLDD41 pKa = 4.02FGPYY45 pKa = 9.37GNKK48 pKa = 10.26LPGDD52 pKa = 3.77GQNYY56 pKa = 9.52CEE58 pKa = 4.25PTSAVMSLFWLGSNGFTQLTPSTYY82 pKa = 10.0PGQNDD87 pKa = 4.27PYY89 pKa = 9.62TYY91 pKa = 10.86NLEE94 pKa = 4.05RR95 pKa = 11.84LMGGLMQTSATQGTYY110 pKa = 10.53DD111 pKa = 3.11SSAIQGISDD120 pKa = 3.56YY121 pKa = 10.97LAAAGLSGSFSYY133 pKa = 10.15IYY135 pKa = 10.69NGGDD139 pKa = 3.27SASNLLNPSWLAAQLAPNAGANPGSITFIDD169 pKa = 5.97FSISWYY175 pKa = 9.04GQNSGSSTSFSAEE188 pKa = 3.89GGHH191 pKa = 6.15EE192 pKa = 4.09LALLAVVDD200 pKa = 4.15PTGNAQLILNNAYY213 pKa = 9.61PSTLEE218 pKa = 3.81NVPNVPEE225 pKa = 4.95SDD227 pKa = 3.68PQTLGITQVPAGWNLPGMALPSEE250 pKa = 5.63DD251 pKa = 3.41YY252 pKa = 10.06WQLQSSDD259 pKa = 3.35GTVGDD264 pKa = 4.08GSTTKK269 pKa = 10.74AIVDD273 pKa = 3.52SAYY276 pKa = 10.57AFSVDD281 pKa = 3.16TMANPTLNPSWTPADD296 pKa = 3.52WVISGSKK303 pKa = 9.15TINTNSATFTVQAAVTGAGGFNKK326 pKa = 9.71TGEE329 pKa = 4.3GALVLTAGDD338 pKa = 4.6DD339 pKa = 3.77LTGNNAVSGGALGSTNAGDD358 pKa = 3.94STPLGTGSFVLTNGGTIGLTPAGSGGPLAFSIASGSGSSVTFGAGGGALEE408 pKa = 4.63IDD410 pKa = 3.16AGGYY414 pKa = 10.37DD415 pKa = 3.86SLSVTMGDD423 pKa = 4.02GSATSSNLIRR433 pKa = 11.84QSAGTFVIIPAHH445 pKa = 6.61GISALGTTEE454 pKa = 3.75RR455 pKa = 11.84VLVAGGSSSRR465 pKa = 11.84PASVNGMVAPYY476 pKa = 10.12IVGQDD481 pKa = 3.22NDD483 pKa = 4.26AGASGYY489 pKa = 9.82FLNYY493 pKa = 9.83DD494 pKa = 3.95GSAGFQKK501 pKa = 10.61AAAVSSLSTAIDD513 pKa = 3.51VATSSTVYY521 pKa = 10.44AAEE524 pKa = 4.11TDD526 pKa = 3.6QVIATNEE533 pKa = 3.96TARR536 pKa = 11.84VYY538 pKa = 10.91ALQVNDD544 pKa = 4.28SEE546 pKa = 5.08VSGTNATLEE555 pKa = 4.23VGSRR559 pKa = 11.84QAGDD563 pKa = 3.35SAGVILNGGTISASSLDD580 pKa = 3.51FGAAEE585 pKa = 4.09GVIYY589 pKa = 10.56ASEE592 pKa = 4.2AGGAISSTITSSAGLTTFGPGTVALSGDD620 pKa = 3.85STSTLSGTVTVNSGTLSAQSAQGSATGSANVSVQANATLSVSGAGAVAGNVSVQNNGTLLLSGGTISGNVTVAEE694 pKa = 4.5VGQSSANPGGTLAGSGTLSGSTTARR719 pKa = 11.84GVITSGASGGNITFEE734 pKa = 4.32GSADD738 pKa = 3.69LNSSQFFWQLDD749 pKa = 3.81SLTDD753 pKa = 3.82DD754 pKa = 4.24ASQAGIEE761 pKa = 4.19WNLLTFLGDD770 pKa = 3.44SHH772 pKa = 8.24VGSEE776 pKa = 4.28DD777 pKa = 3.71APVTIYY783 pKa = 10.9LQFAAGVITPDD794 pKa = 3.19TADD797 pKa = 4.11PFWMTDD803 pKa = 3.63HH804 pKa = 6.72EE805 pKa = 4.53WKK807 pKa = 10.2IWDD810 pKa = 4.1FSQATSYY817 pKa = 10.93SWDD820 pKa = 3.49DD821 pKa = 3.69SGDD824 pKa = 3.52GNTSFNYY831 pKa = 10.43GNFDD835 pKa = 3.83VVEE838 pKa = 4.27KK839 pKa = 10.76AADD842 pKa = 3.28NSVYY846 pKa = 10.43LKK848 pKa = 10.55YY849 pKa = 10.84VAVPEE854 pKa = 4.24PAVIWLALAGCGAMIASRR872 pKa = 11.84RR873 pKa = 11.84IRR875 pKa = 11.84QRR877 pKa = 11.84RR878 pKa = 3.26
Molecular weight: 88.22 kDa
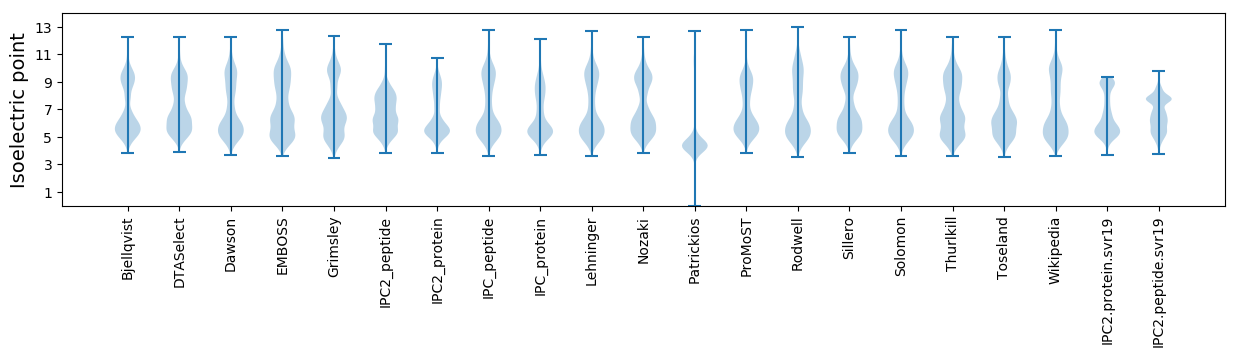
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A146G4Z7|A0A146G4Z7_TERSA LacI family transcriptional regulator OS=Terrimicrobium sacchariphilum OX=690879 GN=TSACC_21070 PE=4 SV=1
MM1 pKa = 7.14LTRR4 pKa = 11.84RR5 pKa = 11.84EE6 pKa = 4.14NGTLPSFTLRR16 pKa = 11.84SDD18 pKa = 3.75TLPLSLRR25 pKa = 11.84HH26 pKa = 6.19AGRR29 pKa = 11.84SGRR32 pKa = 11.84GEE34 pKa = 4.05RR35 pKa = 11.84HH36 pKa = 5.72EE37 pKa = 4.2MTPPRR42 pKa = 11.84AGLWFRR48 pKa = 11.84ILAVEE53 pKa = 4.62TWQLVLLDD61 pKa = 4.74LYY63 pKa = 10.38IAAWAIGMVLQTLLVRR79 pKa = 11.84WLKK82 pKa = 11.07AKK84 pKa = 9.81TSEE87 pKa = 4.5EE88 pKa = 4.35LDD90 pKa = 3.37PHH92 pKa = 6.93LGPGARR98 pKa = 11.84AKK100 pKa = 10.88LSGYY104 pKa = 10.14SIASSMILLRR114 pKa = 11.84LIFTDD119 pKa = 3.94TPLSRR124 pKa = 11.84SPDD127 pKa = 3.28LRR129 pKa = 11.84TICLWLRR136 pKa = 11.84GCVILIAVSLAAFPFLMFLSS156 pKa = 4.09
MM1 pKa = 7.14LTRR4 pKa = 11.84RR5 pKa = 11.84EE6 pKa = 4.14NGTLPSFTLRR16 pKa = 11.84SDD18 pKa = 3.75TLPLSLRR25 pKa = 11.84HH26 pKa = 6.19AGRR29 pKa = 11.84SGRR32 pKa = 11.84GEE34 pKa = 4.05RR35 pKa = 11.84HH36 pKa = 5.72EE37 pKa = 4.2MTPPRR42 pKa = 11.84AGLWFRR48 pKa = 11.84ILAVEE53 pKa = 4.62TWQLVLLDD61 pKa = 4.74LYY63 pKa = 10.38IAAWAIGMVLQTLLVRR79 pKa = 11.84WLKK82 pKa = 11.07AKK84 pKa = 9.81TSEE87 pKa = 4.5EE88 pKa = 4.35LDD90 pKa = 3.37PHH92 pKa = 6.93LGPGARR98 pKa = 11.84AKK100 pKa = 10.88LSGYY104 pKa = 10.14SIASSMILLRR114 pKa = 11.84LIFTDD119 pKa = 3.94TPLSRR124 pKa = 11.84SPDD127 pKa = 3.28LRR129 pKa = 11.84TICLWLRR136 pKa = 11.84GCVILIAVSLAAFPFLMFLSS156 pKa = 4.09
Molecular weight: 17.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1458384 |
25 |
3801 |
349.9 |
38.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.45 ± 0.042 | 0.973 ± 0.014 |
5.186 ± 0.026 | 5.815 ± 0.042 |
4.146 ± 0.024 | 8.221 ± 0.049 |
1.975 ± 0.021 | 5.378 ± 0.03 |
3.877 ± 0.034 | 9.981 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.071 ± 0.015 | 3.202 ± 0.031 |
5.356 ± 0.029 | 3.369 ± 0.022 |
6.407 ± 0.036 | 6.487 ± 0.035 |
5.681 ± 0.038 | 7.129 ± 0.034 |
1.621 ± 0.017 | 2.676 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
