
Tortispora caseinolytica NRRL Y-17796
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Trigonopsidaceae; Tortispora; Tortispora caseinolytica
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
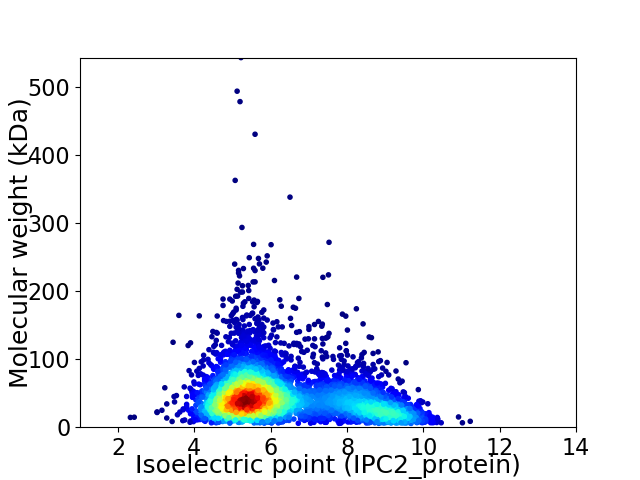
Virtual 2D-PAGE plot for 4657 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E4T9R0|A0A1E4T9R0_9ASCO Uncharacterized protein OS=Tortispora caseinolytica NRRL Y-17796 OX=767744 GN=CANCADRAFT_46371 PE=4 SV=1
MM1 pKa = 7.68LSTVTLTEE9 pKa = 4.03TSLCKK14 pKa = 10.25PPTTTTTVYY23 pKa = 10.48EE24 pKa = 4.26KK25 pKa = 9.97TYY27 pKa = 10.79SSANGIVTAYY37 pKa = 10.62DD38 pKa = 4.26DD39 pKa = 3.94YY40 pKa = 11.61CLPEE44 pKa = 4.23TKK46 pKa = 10.29FVDD49 pKa = 3.45NVYY52 pKa = 10.57CYY54 pKa = 11.15ACTPTVLTYY63 pKa = 10.52VPKK66 pKa = 10.83GPLVNDD72 pKa = 2.94SHH74 pKa = 6.38TYY76 pKa = 9.42VWGLYY81 pKa = 8.05QEE83 pKa = 4.68ISFSVSIAFDD93 pKa = 3.15ITIEE97 pKa = 4.12KK98 pKa = 9.39PIAVTKK104 pKa = 10.28FEE106 pKa = 4.37NADD109 pKa = 4.02AIQTTLSSVALMPVIPLSSNQASISSEE136 pKa = 3.33ISTSPSSILATTSTFSNISVGEE158 pKa = 3.86GAIIGVASATAVEE171 pKa = 4.51STTVQEE177 pKa = 4.66RR178 pKa = 11.84PTAYY182 pKa = 8.4ITSNDD187 pKa = 3.13IRR189 pKa = 11.84NGDD192 pKa = 3.42LTTIVEE198 pKa = 4.59SFKK201 pKa = 11.24EE202 pKa = 4.05MAVSSSVEE210 pKa = 3.58ISSFAQLSSSYY221 pKa = 11.04AEE223 pKa = 4.14SSPALTSLISSSVVQNDD240 pKa = 2.88ISSSYY245 pKa = 10.29PPTSIPKK252 pKa = 9.34VDD254 pKa = 4.06SSSMDD259 pKa = 3.35AVIVAPEE266 pKa = 3.97YY267 pKa = 11.26SLDD270 pKa = 4.06PGPDD274 pKa = 3.72RR275 pKa = 11.84IPQSEE280 pKa = 4.41MVDD283 pKa = 4.23DD284 pKa = 5.64IIKK287 pKa = 11.03SMIDD291 pKa = 3.17GLIIATHH298 pKa = 6.37LPVVSIDD305 pKa = 3.68PGEE308 pKa = 4.76HH309 pKa = 6.57NDD311 pKa = 3.66DD312 pKa = 3.91TVSDD316 pKa = 4.07GLVGDD321 pKa = 4.6NEE323 pKa = 4.94LDD325 pKa = 3.37TGIVDD330 pKa = 3.96SSEE333 pKa = 4.06DD334 pKa = 3.94EE335 pKa = 4.1SWSDD339 pKa = 3.24IVQDD343 pKa = 3.5VDD345 pKa = 4.8DD346 pKa = 4.94EE347 pKa = 4.64EE348 pKa = 5.32SVIDD352 pKa = 5.54LEE354 pKa = 5.03NPDD357 pKa = 5.13LLGDD361 pKa = 4.52DD362 pKa = 4.81YY363 pKa = 12.08AEE365 pKa = 4.12EE366 pKa = 4.07WSDD369 pKa = 4.04LNEE372 pKa = 5.17DD373 pKa = 3.57GSAVDD378 pKa = 4.87GVVSTDD384 pKa = 3.81DD385 pKa = 4.38LVPDD389 pKa = 3.62TTGIAEE395 pKa = 4.16TDD397 pKa = 3.77FGSEE401 pKa = 3.66SGTNISGLVGEE412 pKa = 4.56DD413 pKa = 3.42MSWTVSRR420 pKa = 11.84LEE422 pKa = 4.82DD423 pKa = 3.51YY424 pKa = 11.4SGDD427 pKa = 3.76TEE429 pKa = 4.36VTIDD433 pKa = 4.29KK434 pKa = 10.7ADD436 pKa = 3.39KK437 pKa = 10.77
MM1 pKa = 7.68LSTVTLTEE9 pKa = 4.03TSLCKK14 pKa = 10.25PPTTTTTVYY23 pKa = 10.48EE24 pKa = 4.26KK25 pKa = 9.97TYY27 pKa = 10.79SSANGIVTAYY37 pKa = 10.62DD38 pKa = 4.26DD39 pKa = 3.94YY40 pKa = 11.61CLPEE44 pKa = 4.23TKK46 pKa = 10.29FVDD49 pKa = 3.45NVYY52 pKa = 10.57CYY54 pKa = 11.15ACTPTVLTYY63 pKa = 10.52VPKK66 pKa = 10.83GPLVNDD72 pKa = 2.94SHH74 pKa = 6.38TYY76 pKa = 9.42VWGLYY81 pKa = 8.05QEE83 pKa = 4.68ISFSVSIAFDD93 pKa = 3.15ITIEE97 pKa = 4.12KK98 pKa = 9.39PIAVTKK104 pKa = 10.28FEE106 pKa = 4.37NADD109 pKa = 4.02AIQTTLSSVALMPVIPLSSNQASISSEE136 pKa = 3.33ISTSPSSILATTSTFSNISVGEE158 pKa = 3.86GAIIGVASATAVEE171 pKa = 4.51STTVQEE177 pKa = 4.66RR178 pKa = 11.84PTAYY182 pKa = 8.4ITSNDD187 pKa = 3.13IRR189 pKa = 11.84NGDD192 pKa = 3.42LTTIVEE198 pKa = 4.59SFKK201 pKa = 11.24EE202 pKa = 4.05MAVSSSVEE210 pKa = 3.58ISSFAQLSSSYY221 pKa = 11.04AEE223 pKa = 4.14SSPALTSLISSSVVQNDD240 pKa = 2.88ISSSYY245 pKa = 10.29PPTSIPKK252 pKa = 9.34VDD254 pKa = 4.06SSSMDD259 pKa = 3.35AVIVAPEE266 pKa = 3.97YY267 pKa = 11.26SLDD270 pKa = 4.06PGPDD274 pKa = 3.72RR275 pKa = 11.84IPQSEE280 pKa = 4.41MVDD283 pKa = 4.23DD284 pKa = 5.64IIKK287 pKa = 11.03SMIDD291 pKa = 3.17GLIIATHH298 pKa = 6.37LPVVSIDD305 pKa = 3.68PGEE308 pKa = 4.76HH309 pKa = 6.57NDD311 pKa = 3.66DD312 pKa = 3.91TVSDD316 pKa = 4.07GLVGDD321 pKa = 4.6NEE323 pKa = 4.94LDD325 pKa = 3.37TGIVDD330 pKa = 3.96SSEE333 pKa = 4.06DD334 pKa = 3.94EE335 pKa = 4.1SWSDD339 pKa = 3.24IVQDD343 pKa = 3.5VDD345 pKa = 4.8DD346 pKa = 4.94EE347 pKa = 4.64EE348 pKa = 5.32SVIDD352 pKa = 5.54LEE354 pKa = 5.03NPDD357 pKa = 5.13LLGDD361 pKa = 4.52DD362 pKa = 4.81YY363 pKa = 12.08AEE365 pKa = 4.12EE366 pKa = 4.07WSDD369 pKa = 4.04LNEE372 pKa = 5.17DD373 pKa = 3.57GSAVDD378 pKa = 4.87GVVSTDD384 pKa = 3.81DD385 pKa = 4.38LVPDD389 pKa = 3.62TTGIAEE395 pKa = 4.16TDD397 pKa = 3.77FGSEE401 pKa = 3.66SGTNISGLVGEE412 pKa = 4.56DD413 pKa = 3.42MSWTVSRR420 pKa = 11.84LEE422 pKa = 4.82DD423 pKa = 3.51YY424 pKa = 11.4SGDD427 pKa = 3.76TEE429 pKa = 4.36VTIDD433 pKa = 4.29KK434 pKa = 10.7ADD436 pKa = 3.39KK437 pKa = 10.77
Molecular weight: 46.47 kDa
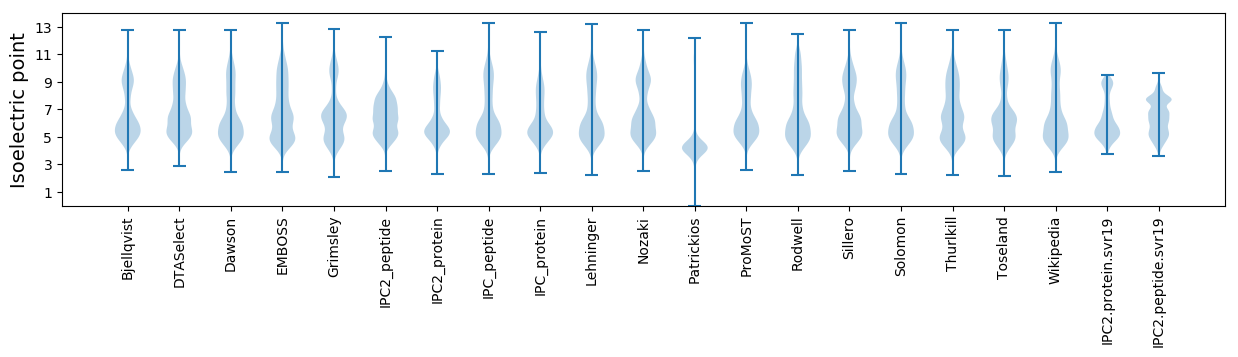
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E4TJU4|A0A1E4TJU4_9ASCO Uncharacterized protein (Fragment) OS=Tortispora caseinolytica NRRL Y-17796 OX=767744 GN=CANCADRAFT_11734 PE=4 SV=1
MM1 pKa = 7.57HH2 pKa = 7.03NQRR5 pKa = 11.84HH6 pKa = 4.78QPSMLQLIMSQPSTFQRR23 pKa = 11.84RR24 pKa = 11.84LKK26 pKa = 10.93SRR28 pKa = 11.84GNTYY32 pKa = 10.22QPSTLRR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 10.01RR41 pKa = 11.84KK42 pKa = 9.22FGFLARR48 pKa = 11.84LRR50 pKa = 11.84SRR52 pKa = 11.84TGKK55 pKa = 10.47KK56 pKa = 9.17ILARR60 pKa = 11.84RR61 pKa = 11.84KK62 pKa = 10.12LKK64 pKa = 10.49GRR66 pKa = 11.84WRR68 pKa = 11.84LTHH71 pKa = 6.58
MM1 pKa = 7.57HH2 pKa = 7.03NQRR5 pKa = 11.84HH6 pKa = 4.78QPSMLQLIMSQPSTFQRR23 pKa = 11.84RR24 pKa = 11.84LKK26 pKa = 10.93SRR28 pKa = 11.84GNTYY32 pKa = 10.22QPSTLRR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 10.01RR41 pKa = 11.84KK42 pKa = 9.22FGFLARR48 pKa = 11.84LRR50 pKa = 11.84SRR52 pKa = 11.84TGKK55 pKa = 10.47KK56 pKa = 9.17ILARR60 pKa = 11.84RR61 pKa = 11.84KK62 pKa = 10.12LKK64 pKa = 10.49GRR66 pKa = 11.84WRR68 pKa = 11.84LTHH71 pKa = 6.58
Molecular weight: 8.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2057799 |
50 |
4853 |
441.9 |
49.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.834 ± 0.03 | 1.312 ± 0.015 |
5.944 ± 0.027 | 6.125 ± 0.032 |
4.032 ± 0.026 | 5.532 ± 0.036 |
2.234 ± 0.017 | 6.048 ± 0.025 |
5.461 ± 0.03 | 9.533 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.149 ± 0.012 | 4.199 ± 0.017 |
5.016 ± 0.03 | 3.643 ± 0.025 |
5.218 ± 0.024 | 9.164 ± 0.062 |
5.667 ± 0.021 | 6.361 ± 0.023 |
1.128 ± 0.012 | 3.399 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
