
Apteryx rowi circovirus-like virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.43
Get precalculated fractions of proteins
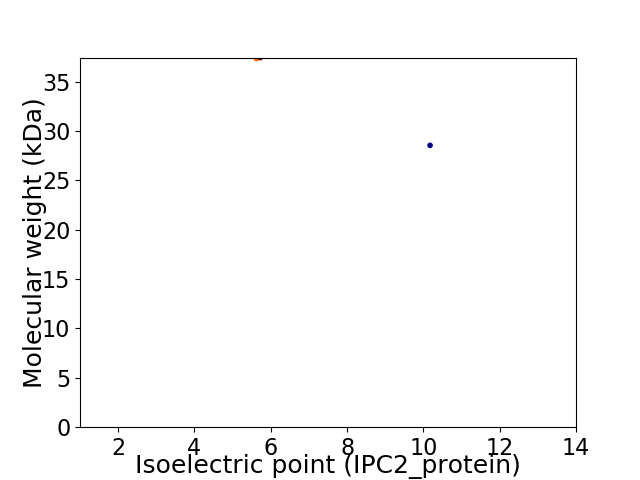
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
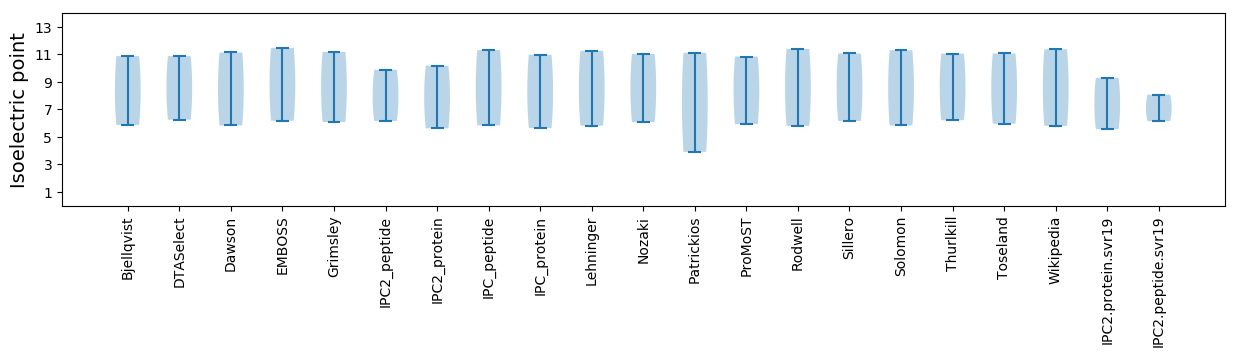
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F6TGK2|A0A0F6TGK2_9CIRC ATP-dependent helicase Rep OS=Apteryx rowi circovirus-like virus OX=1634476 PE=3 SV=1
MM1 pKa = 7.4SKK3 pKa = 10.33ARR5 pKa = 11.84HH6 pKa = 4.78WCFTLNNPDD15 pKa = 4.87DD16 pKa = 4.65EE17 pKa = 5.95PDD19 pKa = 3.6LSDD22 pKa = 4.08TNIQYY27 pKa = 10.78AIYY30 pKa = 9.98QMEE33 pKa = 4.65IAPHH37 pKa = 6.31TDD39 pKa = 2.92TPHH42 pKa = 5.22FQGYY46 pKa = 10.01IILFKK51 pKa = 10.35KK52 pKa = 9.91QRR54 pKa = 11.84LAWVRR59 pKa = 11.84AVLPRR64 pKa = 11.84AHH66 pKa = 6.74WEE68 pKa = 3.87IARR71 pKa = 11.84GTPSQNRR78 pKa = 11.84DD79 pKa = 3.19YY80 pKa = 10.25CTKK83 pKa = 8.72PDD85 pKa = 3.52TRR87 pKa = 11.84IGEE90 pKa = 4.1PSEE93 pKa = 5.3LGLFPEE99 pKa = 5.31DD100 pKa = 3.48AGQGSRR106 pKa = 11.84TDD108 pKa = 3.38LQAVHH113 pKa = 6.93FALQHH118 pKa = 5.55GLTNKK123 pKa = 9.89EE124 pKa = 3.61YY125 pKa = 9.9ANQYY129 pKa = 7.72FSTWIRR135 pKa = 11.84YY136 pKa = 7.57PNLIDD141 pKa = 4.18TFAQSNIVPRR151 pKa = 11.84TGDD154 pKa = 3.49EE155 pKa = 4.26EE156 pKa = 4.45IEE158 pKa = 4.65CILIIGPPGTGKK170 pKa = 10.02SRR172 pKa = 11.84YY173 pKa = 9.05AEE175 pKa = 3.85RR176 pKa = 11.84LARR179 pKa = 11.84SYY181 pKa = 11.55NVGEE185 pKa = 4.42FFRR188 pKa = 11.84KK189 pKa = 9.28QRR191 pKa = 11.84GKK193 pKa = 9.83WYY195 pKa = 10.0DD196 pKa = 3.13GYY198 pKa = 11.04LGEE201 pKa = 5.02RR202 pKa = 11.84VIIFDD207 pKa = 4.08DD208 pKa = 4.06FRR210 pKa = 11.84GSSLSFTDD218 pKa = 5.04FKK220 pKa = 11.59LLVDD224 pKa = 4.69RR225 pKa = 11.84YY226 pKa = 9.8ALRR229 pKa = 11.84VEE231 pKa = 4.33VKK233 pKa = 10.24GRR235 pKa = 11.84YY236 pKa = 8.75VNMAATKK243 pKa = 9.74FIITTNLSPDD253 pKa = 3.23AWWGEE258 pKa = 4.24EE259 pKa = 3.88VTGPEE264 pKa = 3.98FPAISRR270 pKa = 11.84RR271 pKa = 11.84ITEE274 pKa = 3.81VLFFPEE280 pKa = 4.67LNSFCRR286 pKa = 11.84FDD288 pKa = 3.52SYY290 pKa = 11.29EE291 pKa = 4.01AYY293 pKa = 10.68LEE295 pKa = 3.93YY296 pKa = 10.74LRR298 pKa = 11.84PQLILPHH305 pKa = 6.55GLLPQAQAQYY315 pKa = 11.25AFQEE319 pKa = 4.39VIYY322 pKa = 10.8
MM1 pKa = 7.4SKK3 pKa = 10.33ARR5 pKa = 11.84HH6 pKa = 4.78WCFTLNNPDD15 pKa = 4.87DD16 pKa = 4.65EE17 pKa = 5.95PDD19 pKa = 3.6LSDD22 pKa = 4.08TNIQYY27 pKa = 10.78AIYY30 pKa = 9.98QMEE33 pKa = 4.65IAPHH37 pKa = 6.31TDD39 pKa = 2.92TPHH42 pKa = 5.22FQGYY46 pKa = 10.01IILFKK51 pKa = 10.35KK52 pKa = 9.91QRR54 pKa = 11.84LAWVRR59 pKa = 11.84AVLPRR64 pKa = 11.84AHH66 pKa = 6.74WEE68 pKa = 3.87IARR71 pKa = 11.84GTPSQNRR78 pKa = 11.84DD79 pKa = 3.19YY80 pKa = 10.25CTKK83 pKa = 8.72PDD85 pKa = 3.52TRR87 pKa = 11.84IGEE90 pKa = 4.1PSEE93 pKa = 5.3LGLFPEE99 pKa = 5.31DD100 pKa = 3.48AGQGSRR106 pKa = 11.84TDD108 pKa = 3.38LQAVHH113 pKa = 6.93FALQHH118 pKa = 5.55GLTNKK123 pKa = 9.89EE124 pKa = 3.61YY125 pKa = 9.9ANQYY129 pKa = 7.72FSTWIRR135 pKa = 11.84YY136 pKa = 7.57PNLIDD141 pKa = 4.18TFAQSNIVPRR151 pKa = 11.84TGDD154 pKa = 3.49EE155 pKa = 4.26EE156 pKa = 4.45IEE158 pKa = 4.65CILIIGPPGTGKK170 pKa = 10.02SRR172 pKa = 11.84YY173 pKa = 9.05AEE175 pKa = 3.85RR176 pKa = 11.84LARR179 pKa = 11.84SYY181 pKa = 11.55NVGEE185 pKa = 4.42FFRR188 pKa = 11.84KK189 pKa = 9.28QRR191 pKa = 11.84GKK193 pKa = 9.83WYY195 pKa = 10.0DD196 pKa = 3.13GYY198 pKa = 11.04LGEE201 pKa = 5.02RR202 pKa = 11.84VIIFDD207 pKa = 4.08DD208 pKa = 4.06FRR210 pKa = 11.84GSSLSFTDD218 pKa = 5.04FKK220 pKa = 11.59LLVDD224 pKa = 4.69RR225 pKa = 11.84YY226 pKa = 9.8ALRR229 pKa = 11.84VEE231 pKa = 4.33VKK233 pKa = 10.24GRR235 pKa = 11.84YY236 pKa = 8.75VNMAATKK243 pKa = 9.74FIITTNLSPDD253 pKa = 3.23AWWGEE258 pKa = 4.24EE259 pKa = 3.88VTGPEE264 pKa = 3.98FPAISRR270 pKa = 11.84RR271 pKa = 11.84ITEE274 pKa = 3.81VLFFPEE280 pKa = 4.67LNSFCRR286 pKa = 11.84FDD288 pKa = 3.52SYY290 pKa = 11.29EE291 pKa = 4.01AYY293 pKa = 10.68LEE295 pKa = 3.93YY296 pKa = 10.74LRR298 pKa = 11.84PQLILPHH305 pKa = 6.55GLLPQAQAQYY315 pKa = 11.25AFQEE319 pKa = 4.39VIYY322 pKa = 10.8
Molecular weight: 37.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F6TGK2|A0A0F6TGK2_9CIRC ATP-dependent helicase Rep OS=Apteryx rowi circovirus-like virus OX=1634476 PE=3 SV=1
MM1 pKa = 7.05VSYY4 pKa = 10.52RR5 pKa = 11.84KK6 pKa = 9.51RR7 pKa = 11.84KK8 pKa = 9.55RR9 pKa = 11.84SMRR12 pKa = 11.84SKK14 pKa = 10.99KK15 pKa = 10.52SFTKK19 pKa = 9.67IKK21 pKa = 10.21RR22 pKa = 11.84RR23 pKa = 11.84PRR25 pKa = 11.84VLNKK29 pKa = 9.72FKK31 pKa = 10.77RR32 pKa = 11.84KK33 pKa = 9.19NRR35 pKa = 11.84GSQRR39 pKa = 11.84IKK41 pKa = 8.39RR42 pKa = 11.84TRR44 pKa = 11.84WPVDD48 pKa = 3.11NPFGDD53 pKa = 3.43NLFYY57 pKa = 10.77KK58 pKa = 10.63VRR60 pKa = 11.84FSYY63 pKa = 8.93GTTLTIPTTASSINLGFQMNSLASIISSAGNAPGLNSLGNAFVNYY108 pKa = 9.96RR109 pKa = 11.84IRR111 pKa = 11.84GLKK114 pKa = 9.12TKK116 pKa = 7.79MTLWPDD122 pKa = 3.18SATVPIVAYY131 pKa = 9.48HH132 pKa = 6.73LAGASQSEE140 pKa = 4.59MASITPNVSTLPEE153 pKa = 4.26LRR155 pKa = 11.84WQKK158 pKa = 10.85YY159 pKa = 5.69RR160 pKa = 11.84TCNYY164 pKa = 9.15AAAGAKK170 pKa = 7.37PTRR173 pKa = 11.84LNSYY177 pKa = 9.44FSVDD181 pKa = 3.21KK182 pKa = 11.18VYY184 pKa = 11.13GPDD187 pKa = 3.13AVVKK191 pKa = 10.58NDD193 pKa = 3.31SNFVGNITTAAPPAPPVFGAPGLGPGFRR221 pKa = 11.84FGLFTMSGLNPTVAVNCVTKK241 pKa = 10.4IEE243 pKa = 4.06FTAYY247 pKa = 10.76VNFFSKK253 pKa = 10.5RR254 pKa = 11.84GIINN258 pKa = 3.34
MM1 pKa = 7.05VSYY4 pKa = 10.52RR5 pKa = 11.84KK6 pKa = 9.51RR7 pKa = 11.84KK8 pKa = 9.55RR9 pKa = 11.84SMRR12 pKa = 11.84SKK14 pKa = 10.99KK15 pKa = 10.52SFTKK19 pKa = 9.67IKK21 pKa = 10.21RR22 pKa = 11.84RR23 pKa = 11.84PRR25 pKa = 11.84VLNKK29 pKa = 9.72FKK31 pKa = 10.77RR32 pKa = 11.84KK33 pKa = 9.19NRR35 pKa = 11.84GSQRR39 pKa = 11.84IKK41 pKa = 8.39RR42 pKa = 11.84TRR44 pKa = 11.84WPVDD48 pKa = 3.11NPFGDD53 pKa = 3.43NLFYY57 pKa = 10.77KK58 pKa = 10.63VRR60 pKa = 11.84FSYY63 pKa = 8.93GTTLTIPTTASSINLGFQMNSLASIISSAGNAPGLNSLGNAFVNYY108 pKa = 9.96RR109 pKa = 11.84IRR111 pKa = 11.84GLKK114 pKa = 9.12TKK116 pKa = 7.79MTLWPDD122 pKa = 3.18SATVPIVAYY131 pKa = 9.48HH132 pKa = 6.73LAGASQSEE140 pKa = 4.59MASITPNVSTLPEE153 pKa = 4.26LRR155 pKa = 11.84WQKK158 pKa = 10.85YY159 pKa = 5.69RR160 pKa = 11.84TCNYY164 pKa = 9.15AAAGAKK170 pKa = 7.37PTRR173 pKa = 11.84LNSYY177 pKa = 9.44FSVDD181 pKa = 3.21KK182 pKa = 11.18VYY184 pKa = 11.13GPDD187 pKa = 3.13AVVKK191 pKa = 10.58NDD193 pKa = 3.31SNFVGNITTAAPPAPPVFGAPGLGPGFRR221 pKa = 11.84FGLFTMSGLNPTVAVNCVTKK241 pKa = 10.4IEE243 pKa = 4.06FTAYY247 pKa = 10.76VNFFSKK253 pKa = 10.5RR254 pKa = 11.84GIINN258 pKa = 3.34
Molecular weight: 28.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
580 |
258 |
322 |
290.0 |
32.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.586 ± 0.347 | 1.034 ± 0.163 |
4.138 ± 1.137 | 4.31 ± 1.974 |
6.207 ± 0.003 | 6.724 ± 0.402 |
1.379 ± 0.622 | 6.034 ± 0.624 |
5.172 ± 1.375 | 7.414 ± 0.76 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.552 ± 0.485 | 5.517 ± 1.402 |
6.552 ± 0.267 | 3.448 ± 1.19 |
7.586 ± 0.104 | 6.724 ± 1.374 |
6.724 ± 0.645 | 5.345 ± 1.023 |
1.724 ± 0.352 | 4.828 ± 0.597 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
