
McMurdo Ice Shelf pond-associated circular DNA virus-1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.83
Get precalculated fractions of proteins
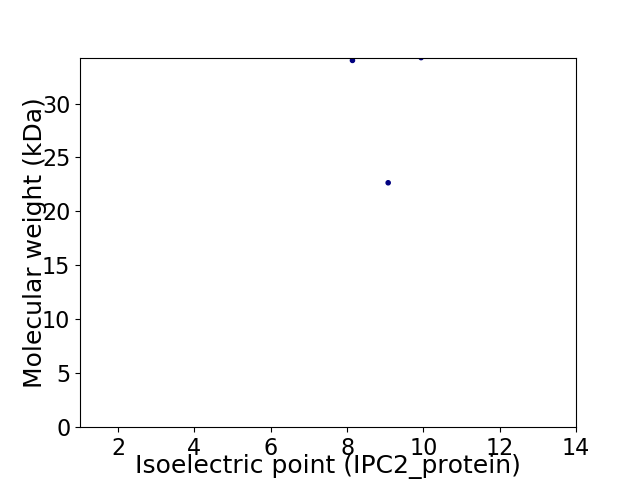
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A075LZD6|A0A075LZD6_9VIRU Uncharacterized protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-1 OX=1521385 PE=4 SV=1
MM1 pKa = 7.27SQGKK5 pKa = 8.11HH6 pKa = 4.22WQFTLNNPTQDD17 pKa = 3.35EE18 pKa = 4.64RR19 pKa = 11.84NVLAEE24 pKa = 5.27LGDD27 pKa = 3.89QPTTQYY33 pKa = 11.14LIYY36 pKa = 9.68GDD38 pKa = 4.13EE39 pKa = 4.39VGASGTPHH47 pKa = 6.34LQGHH51 pKa = 5.35VSFVQRR57 pKa = 11.84YY58 pKa = 8.4RR59 pKa = 11.84FNQVKK64 pKa = 9.93NWVSPRR70 pKa = 11.84AHH72 pKa = 6.65LEE74 pKa = 3.67LVRR77 pKa = 11.84LLRR80 pKa = 11.84RR81 pKa = 11.84HH82 pKa = 6.28IEE84 pKa = 3.93YY85 pKa = 10.32CKK87 pKa = 10.53KK88 pKa = 10.54DD89 pKa = 3.25GAYY92 pKa = 10.66LEE94 pKa = 5.39FGTPPDD100 pKa = 3.36STLAKK105 pKa = 10.15DD106 pKa = 4.01GKK108 pKa = 10.44RR109 pKa = 11.84NEE111 pKa = 4.15LAEE114 pKa = 4.12FRR116 pKa = 11.84ATVAEE121 pKa = 4.77GVFHH125 pKa = 7.27SPEE128 pKa = 3.93LRR130 pKa = 11.84EE131 pKa = 3.75KK132 pKa = 10.35HH133 pKa = 6.16PNVMARR139 pKa = 11.84YY140 pKa = 7.83PHH142 pKa = 6.62FANSIIRR149 pKa = 11.84DD150 pKa = 4.45LFPQSAPPDD159 pKa = 3.58LPLRR163 pKa = 11.84AWQQRR168 pKa = 11.84VVEE171 pKa = 5.37LIDD174 pKa = 4.6APPDD178 pKa = 3.34PRR180 pKa = 11.84KK181 pKa = 10.53VYY183 pKa = 10.19FIVDD187 pKa = 3.53RR188 pKa = 11.84QGNAGKK194 pKa = 8.76TSLAKK199 pKa = 10.38LLHH202 pKa = 5.93RR203 pKa = 11.84THH205 pKa = 6.7EE206 pKa = 4.21AVQIIRR212 pKa = 11.84SGKK215 pKa = 7.81VADD218 pKa = 3.49MAYY221 pKa = 10.33LYY223 pKa = 10.68KK224 pKa = 9.69ITTKK228 pKa = 10.48ILILDD233 pKa = 3.84VPRR236 pKa = 11.84SKK238 pKa = 11.17SEE240 pKa = 3.62LLQYY244 pKa = 11.17SFIEE248 pKa = 4.16MVKK251 pKa = 10.6DD252 pKa = 3.54GLLMSTKK259 pKa = 10.05YY260 pKa = 10.29EE261 pKa = 4.22SVMKK265 pKa = 10.38TFDD268 pKa = 3.76PPHH271 pKa = 7.68ILVMMNADD279 pKa = 4.04PDD281 pKa = 3.9HH282 pKa = 6.7TALSTDD288 pKa = 3.47RR289 pKa = 11.84YY290 pKa = 10.54HH291 pKa = 7.72YY292 pKa = 10.0IIPHH296 pKa = 6.01
MM1 pKa = 7.27SQGKK5 pKa = 8.11HH6 pKa = 4.22WQFTLNNPTQDD17 pKa = 3.35EE18 pKa = 4.64RR19 pKa = 11.84NVLAEE24 pKa = 5.27LGDD27 pKa = 3.89QPTTQYY33 pKa = 11.14LIYY36 pKa = 9.68GDD38 pKa = 4.13EE39 pKa = 4.39VGASGTPHH47 pKa = 6.34LQGHH51 pKa = 5.35VSFVQRR57 pKa = 11.84YY58 pKa = 8.4RR59 pKa = 11.84FNQVKK64 pKa = 9.93NWVSPRR70 pKa = 11.84AHH72 pKa = 6.65LEE74 pKa = 3.67LVRR77 pKa = 11.84LLRR80 pKa = 11.84RR81 pKa = 11.84HH82 pKa = 6.28IEE84 pKa = 3.93YY85 pKa = 10.32CKK87 pKa = 10.53KK88 pKa = 10.54DD89 pKa = 3.25GAYY92 pKa = 10.66LEE94 pKa = 5.39FGTPPDD100 pKa = 3.36STLAKK105 pKa = 10.15DD106 pKa = 4.01GKK108 pKa = 10.44RR109 pKa = 11.84NEE111 pKa = 4.15LAEE114 pKa = 4.12FRR116 pKa = 11.84ATVAEE121 pKa = 4.77GVFHH125 pKa = 7.27SPEE128 pKa = 3.93LRR130 pKa = 11.84EE131 pKa = 3.75KK132 pKa = 10.35HH133 pKa = 6.16PNVMARR139 pKa = 11.84YY140 pKa = 7.83PHH142 pKa = 6.62FANSIIRR149 pKa = 11.84DD150 pKa = 4.45LFPQSAPPDD159 pKa = 3.58LPLRR163 pKa = 11.84AWQQRR168 pKa = 11.84VVEE171 pKa = 5.37LIDD174 pKa = 4.6APPDD178 pKa = 3.34PRR180 pKa = 11.84KK181 pKa = 10.53VYY183 pKa = 10.19FIVDD187 pKa = 3.53RR188 pKa = 11.84QGNAGKK194 pKa = 8.76TSLAKK199 pKa = 10.38LLHH202 pKa = 5.93RR203 pKa = 11.84THH205 pKa = 6.7EE206 pKa = 4.21AVQIIRR212 pKa = 11.84SGKK215 pKa = 7.81VADD218 pKa = 3.49MAYY221 pKa = 10.33LYY223 pKa = 10.68KK224 pKa = 9.69ITTKK228 pKa = 10.48ILILDD233 pKa = 3.84VPRR236 pKa = 11.84SKK238 pKa = 11.17SEE240 pKa = 3.62LLQYY244 pKa = 11.17SFIEE248 pKa = 4.16MVKK251 pKa = 10.6DD252 pKa = 3.54GLLMSTKK259 pKa = 10.05YY260 pKa = 10.29EE261 pKa = 4.22SVMKK265 pKa = 10.38TFDD268 pKa = 3.76PPHH271 pKa = 7.68ILVMMNADD279 pKa = 4.04PDD281 pKa = 3.9HH282 pKa = 6.7TALSTDD288 pKa = 3.47RR289 pKa = 11.84YY290 pKa = 10.54HH291 pKa = 7.72YY292 pKa = 10.0IIPHH296 pKa = 6.01
Molecular weight: 34.0 kDa
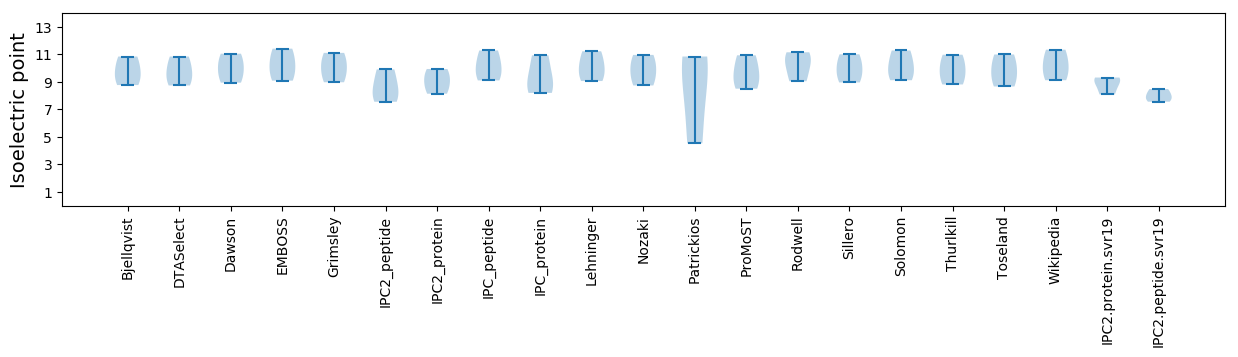
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075LZ71|A0A075LZ71_9VIRU Replication-associated protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-1 OX=1521385 PE=4 SV=1
MM1 pKa = 7.64PKK3 pKa = 9.92VRR5 pKa = 11.84WDD7 pKa = 4.07DD8 pKa = 3.58VVKK11 pKa = 10.36WGIFMPWGRR20 pKa = 11.84SSLMRR25 pKa = 11.84HH26 pKa = 5.61WDD28 pKa = 3.45NDD30 pKa = 3.36YY31 pKa = 11.2KK32 pKa = 10.99QYY34 pKa = 11.24NDD36 pKa = 4.28PKK38 pKa = 9.52TIQYY42 pKa = 10.73KK43 pKa = 10.06DD44 pKa = 2.83GSMAKK49 pKa = 9.63RR50 pKa = 11.84KK51 pKa = 9.25SASVTVFGGQASSKK65 pKa = 9.46RR66 pKa = 11.84ARR68 pKa = 11.84TRR70 pKa = 11.84SMGRR74 pKa = 11.84IRR76 pKa = 11.84RR77 pKa = 11.84VTRR80 pKa = 11.84YY81 pKa = 9.22RR82 pKa = 11.84RR83 pKa = 11.84GRR85 pKa = 11.84RR86 pKa = 11.84GASRR90 pKa = 11.84KK91 pKa = 8.37RR92 pKa = 11.84TSSYY96 pKa = 9.44GARR99 pKa = 11.84ARR101 pKa = 11.84RR102 pKa = 11.84QIGEE106 pKa = 4.04RR107 pKa = 11.84VGTHH111 pKa = 4.24SAKK114 pKa = 10.15TKK116 pKa = 9.54VVEE119 pKa = 4.48DD120 pKa = 3.81AVTTFAMSTRR130 pKa = 11.84TLYY133 pKa = 11.07NLVVSEE139 pKa = 4.11VLEE142 pKa = 4.15GTEE145 pKa = 3.82RR146 pKa = 11.84NEE148 pKa = 4.28RR149 pKa = 11.84VNDD152 pKa = 3.56KK153 pKa = 11.23VNFRR157 pKa = 11.84GIKK160 pKa = 10.23VFLQAYY166 pKa = 7.49NALNTPVNFHH176 pKa = 5.86VAYY179 pKa = 10.5LSPKK183 pKa = 9.44VGQTIDD189 pKa = 3.19TTDD192 pKa = 3.89FFRR195 pKa = 11.84SEE197 pKa = 4.13RR198 pKa = 11.84IARR201 pKa = 11.84GRR203 pKa = 11.84NFATIEE209 pKa = 3.98LSSLEE214 pKa = 4.13MRR216 pKa = 11.84TLPINTDD223 pKa = 3.0RR224 pKa = 11.84YY225 pKa = 10.39VVLKK229 pKa = 9.92HH230 pKa = 4.89MRR232 pKa = 11.84MRR234 pKa = 11.84LAAATEE240 pKa = 3.85ASTIFEE246 pKa = 4.78DD247 pKa = 4.03GGRR250 pKa = 11.84PSLMTAQFYY259 pKa = 11.1LPIKK263 pKa = 9.22RR264 pKa = 11.84QLRR267 pKa = 11.84FEE269 pKa = 4.74GPGSDD274 pKa = 3.42TPIDD278 pKa = 4.18GKK280 pKa = 9.45SLWFIGATFLQQMLVLRR297 pKa = 11.84YY298 pKa = 9.36
MM1 pKa = 7.64PKK3 pKa = 9.92VRR5 pKa = 11.84WDD7 pKa = 4.07DD8 pKa = 3.58VVKK11 pKa = 10.36WGIFMPWGRR20 pKa = 11.84SSLMRR25 pKa = 11.84HH26 pKa = 5.61WDD28 pKa = 3.45NDD30 pKa = 3.36YY31 pKa = 11.2KK32 pKa = 10.99QYY34 pKa = 11.24NDD36 pKa = 4.28PKK38 pKa = 9.52TIQYY42 pKa = 10.73KK43 pKa = 10.06DD44 pKa = 2.83GSMAKK49 pKa = 9.63RR50 pKa = 11.84KK51 pKa = 9.25SASVTVFGGQASSKK65 pKa = 9.46RR66 pKa = 11.84ARR68 pKa = 11.84TRR70 pKa = 11.84SMGRR74 pKa = 11.84IRR76 pKa = 11.84RR77 pKa = 11.84VTRR80 pKa = 11.84YY81 pKa = 9.22RR82 pKa = 11.84RR83 pKa = 11.84GRR85 pKa = 11.84RR86 pKa = 11.84GASRR90 pKa = 11.84KK91 pKa = 8.37RR92 pKa = 11.84TSSYY96 pKa = 9.44GARR99 pKa = 11.84ARR101 pKa = 11.84RR102 pKa = 11.84QIGEE106 pKa = 4.04RR107 pKa = 11.84VGTHH111 pKa = 4.24SAKK114 pKa = 10.15TKK116 pKa = 9.54VVEE119 pKa = 4.48DD120 pKa = 3.81AVTTFAMSTRR130 pKa = 11.84TLYY133 pKa = 11.07NLVVSEE139 pKa = 4.11VLEE142 pKa = 4.15GTEE145 pKa = 3.82RR146 pKa = 11.84NEE148 pKa = 4.28RR149 pKa = 11.84VNDD152 pKa = 3.56KK153 pKa = 11.23VNFRR157 pKa = 11.84GIKK160 pKa = 10.23VFLQAYY166 pKa = 7.49NALNTPVNFHH176 pKa = 5.86VAYY179 pKa = 10.5LSPKK183 pKa = 9.44VGQTIDD189 pKa = 3.19TTDD192 pKa = 3.89FFRR195 pKa = 11.84SEE197 pKa = 4.13RR198 pKa = 11.84IARR201 pKa = 11.84GRR203 pKa = 11.84NFATIEE209 pKa = 3.98LSSLEE214 pKa = 4.13MRR216 pKa = 11.84TLPINTDD223 pKa = 3.0RR224 pKa = 11.84YY225 pKa = 10.39VVLKK229 pKa = 9.92HH230 pKa = 4.89MRR232 pKa = 11.84MRR234 pKa = 11.84LAAATEE240 pKa = 3.85ASTIFEE246 pKa = 4.78DD247 pKa = 4.03GGRR250 pKa = 11.84PSLMTAQFYY259 pKa = 11.1LPIKK263 pKa = 9.22RR264 pKa = 11.84QLRR267 pKa = 11.84FEE269 pKa = 4.74GPGSDD274 pKa = 3.42TPIDD278 pKa = 4.18GKK280 pKa = 9.45SLWFIGATFLQQMLVLRR297 pKa = 11.84YY298 pKa = 9.36
Molecular weight: 34.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
793 |
199 |
298 |
264.3 |
30.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.305 ± 0.66 | 0.631 ± 0.516 |
4.414 ± 1.105 | 4.666 ± 0.349 |
4.035 ± 0.318 | 5.044 ± 1.118 |
2.774 ± 0.887 | 5.17 ± 0.537 |
6.053 ± 0.36 | 9.458 ± 1.768 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.279 ± 0.26 | 3.909 ± 0.413 |
5.422 ± 0.978 | 3.405 ± 0.763 |
8.323 ± 2.037 | 7.945 ± 1.742 |
6.81 ± 0.669 | 6.81 ± 0.332 |
1.765 ± 0.48 | 3.783 ± 0.325 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
