
Beihai sobemo-like virus 21
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
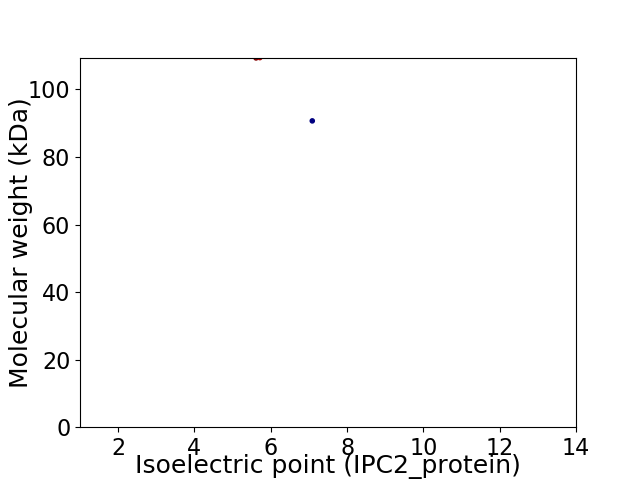
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
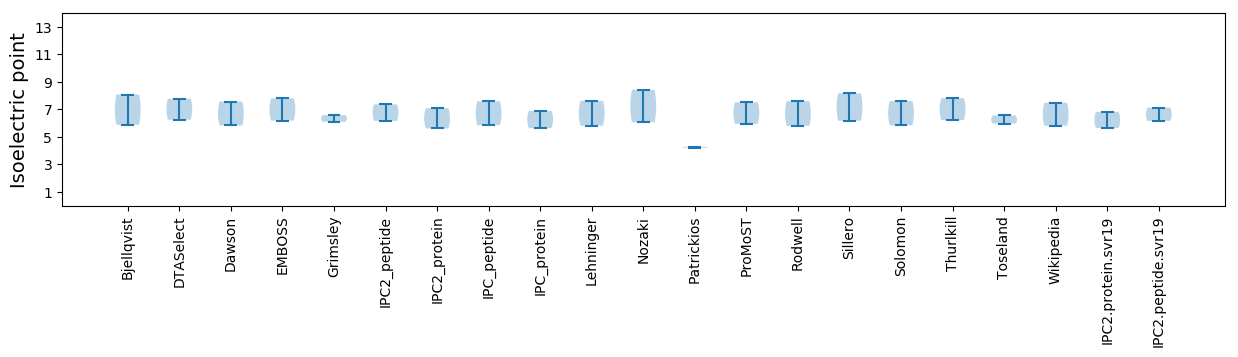
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE94|A0A1L3KE94_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 21 OX=1922693 PE=4 SV=1
MM1 pKa = 7.7RR2 pKa = 11.84LCVSPFMVSCAIIIAVYY19 pKa = 6.83THH21 pKa = 6.61IGIMIGVMVGDD32 pKa = 4.15FTDD35 pKa = 3.83LPFRR39 pKa = 11.84EE40 pKa = 4.52LNKK43 pKa = 10.13QLLRR47 pKa = 11.84THH49 pKa = 6.01YY50 pKa = 10.6VHH52 pKa = 7.39AFWASLNFLGEE63 pKa = 4.53FLRR66 pKa = 11.84SMHH69 pKa = 5.89TFLWALVEE77 pKa = 5.94FILAQNFSLEE87 pKa = 4.37SVGFIALLVVPTIHH101 pKa = 5.7VLYY104 pKa = 10.84VIFRR108 pKa = 11.84PKK110 pKa = 10.09PIRR113 pKa = 11.84FGEE116 pKa = 4.01IEE118 pKa = 4.63EE119 pKa = 4.37ILDD122 pKa = 3.98GYY124 pKa = 11.0LAFTGEE130 pKa = 4.42SLQEE134 pKa = 4.0GSPLQQVQPEE144 pKa = 4.44SVPKK148 pKa = 10.14GQAVLIAEE156 pKa = 4.52TATARR161 pKa = 11.84YY162 pKa = 8.55LQGQVTRR169 pKa = 11.84MDD171 pKa = 3.59NVEE174 pKa = 4.0VTDD177 pKa = 3.87TVIEE181 pKa = 4.14RR182 pKa = 11.84WNKK185 pKa = 10.45LGIDD189 pKa = 3.66YY190 pKa = 10.72DD191 pKa = 4.2FSIVNSGEE199 pKa = 4.26TKK201 pKa = 9.11FACYY205 pKa = 9.97AGLSHH210 pKa = 6.74CTTRR214 pKa = 11.84SGKK217 pKa = 9.93YY218 pKa = 10.04FIAKK222 pKa = 10.29GNDD225 pKa = 3.15LNVQVPCYY233 pKa = 10.03SVPSLAAEE241 pKa = 3.96DD242 pKa = 4.4LMFFLVPANKK252 pKa = 9.76VSVLGNPVVKK262 pKa = 9.93PHH264 pKa = 6.15STDD267 pKa = 2.85LPANVRR273 pKa = 11.84VITSAAKK280 pKa = 9.4YY281 pKa = 10.05GEE283 pKa = 4.5KK284 pKa = 10.19FGSWSTFGRR293 pKa = 11.84LDD295 pKa = 3.68RR296 pKa = 11.84DD297 pKa = 4.12DD298 pKa = 3.68VTGCCRR304 pKa = 11.84FTGSTISGYY313 pKa = 10.78SGAGYY318 pKa = 9.49YY319 pKa = 10.57ASNAWVGIHH328 pKa = 6.53RR329 pKa = 11.84GHH331 pKa = 7.37RR332 pKa = 11.84SGLNFGFLAAYY343 pKa = 9.38VKK345 pKa = 9.27TVKK348 pKa = 10.55NVTFLDD354 pKa = 3.71KK355 pKa = 11.01AVTGEE360 pKa = 4.17SDD362 pKa = 3.43TEE364 pKa = 4.05LARR367 pKa = 11.84LLGLLRR373 pKa = 11.84EE374 pKa = 5.04DD375 pKa = 4.55GPMAYY380 pKa = 10.12SKK382 pKa = 10.77LSKK385 pKa = 10.15RR386 pKa = 11.84KK387 pKa = 10.27SGDD390 pKa = 3.24PSIDD394 pKa = 3.2LYY396 pKa = 11.03EE397 pKa = 4.05YY398 pKa = 10.74NGDD401 pKa = 3.47YY402 pKa = 10.63FYY404 pKa = 11.76VDD406 pKa = 3.36RR407 pKa = 11.84DD408 pKa = 3.99DD409 pKa = 3.94VNSYY413 pKa = 8.44QRR415 pKa = 11.84EE416 pKa = 3.75EE417 pKa = 3.85WQNYY421 pKa = 7.89LDD423 pKa = 4.87AMQMDD428 pKa = 4.8NEE430 pKa = 4.44TEE432 pKa = 3.84DD433 pKa = 3.49RR434 pKa = 11.84QVRR437 pKa = 11.84RR438 pKa = 11.84RR439 pKa = 11.84NKK441 pKa = 9.53SRR443 pKa = 11.84RR444 pKa = 11.84RR445 pKa = 11.84DD446 pKa = 3.49DD447 pKa = 3.41TDD449 pKa = 3.76DD450 pKa = 4.17FEE452 pKa = 7.14GEE454 pKa = 4.09AGSVDD459 pKa = 3.87LTHH462 pKa = 6.84LNSRR466 pKa = 11.84GGAVRR471 pKa = 11.84TEE473 pKa = 4.28CGEE476 pKa = 4.2EE477 pKa = 3.76IQGTMLLPPPRR488 pKa = 11.84CDD490 pKa = 2.86STGNVTDD497 pKa = 3.57HH498 pKa = 6.71ARR500 pKa = 11.84EE501 pKa = 3.8KK502 pKa = 10.72RR503 pKa = 11.84ADD505 pKa = 4.17EE506 pKa = 4.1IALLLIKK513 pKa = 10.53QKK515 pKa = 10.83LDD517 pKa = 3.34RR518 pKa = 11.84LSEE521 pKa = 4.01KK522 pKa = 9.91MKK524 pKa = 10.74QSFVNYY530 pKa = 9.11TFEE533 pKa = 5.17QKK535 pKa = 10.49LGWMKK540 pKa = 10.55SPQEE544 pKa = 3.84LWHH547 pKa = 6.27LTGKK551 pKa = 8.82MLSIQSALTHH561 pKa = 6.5WIKK564 pKa = 10.56PVAEE568 pKa = 4.85AYY570 pKa = 10.28SEE572 pKa = 4.25TMSRR576 pKa = 11.84VAEE579 pKa = 3.91LTPWEE584 pKa = 4.13EE585 pKa = 3.8EE586 pKa = 4.13AKK588 pKa = 10.63KK589 pKa = 10.67KK590 pKa = 9.58LAKK593 pKa = 10.25SRR595 pKa = 11.84ASYY598 pKa = 10.94NKK600 pKa = 10.53ANGTPAPPASFDD612 pKa = 4.61PILEE616 pKa = 4.08SWYY619 pKa = 9.83EE620 pKa = 3.9ATEE623 pKa = 4.01KK624 pKa = 10.61LHH626 pKa = 5.13EE627 pKa = 4.47QIMEE631 pKa = 3.69ISEE634 pKa = 4.58RR635 pKa = 11.84YY636 pKa = 8.24PPPSVTTEE644 pKa = 3.9GDD646 pKa = 3.44VADD649 pKa = 4.69PEE651 pKa = 4.54DD652 pKa = 4.39FEE654 pKa = 6.48GEE656 pKa = 4.33TGDD659 pKa = 4.46HH660 pKa = 5.69QLEE663 pKa = 4.38DD664 pKa = 3.65LLEE667 pKa = 4.18ILRR670 pKa = 11.84NPTQPPLTQPLEE682 pKa = 4.17KK683 pKa = 10.1EE684 pKa = 3.95QTSRR688 pKa = 11.84FQMKK692 pKa = 9.28HH693 pKa = 5.61LSGAPIVKK701 pKa = 8.21QTAAKK706 pKa = 10.44SPTTLPVVPAVISSGDD722 pKa = 3.79DD723 pKa = 3.5LEE725 pKa = 5.08VGADD729 pKa = 3.48AEE731 pKa = 4.63SFSASQAVSHH741 pKa = 6.09PTEE744 pKa = 4.34DD745 pKa = 3.93FIEE748 pKa = 4.72RR749 pKa = 11.84SSLTSQATSRR759 pKa = 11.84PPVLSDD765 pKa = 3.31RR766 pKa = 11.84GQRR769 pKa = 11.84HH770 pKa = 5.82ASASNQLEE778 pKa = 4.3TPLASNGQIQNDD790 pKa = 3.82QPTQLPKK797 pKa = 10.68ASAIIPEE804 pKa = 4.69CSSEE808 pKa = 4.25TTSQMSGPHH817 pKa = 5.81GMNLTKK823 pKa = 10.37SAKK826 pKa = 9.14TSGKK830 pKa = 10.23SSLSSPRR837 pKa = 11.84QEE839 pKa = 4.44SRR841 pKa = 11.84FEE843 pKa = 4.29TVWPEE848 pKa = 3.76KK849 pKa = 10.95SKK851 pKa = 9.82LTSNKK856 pKa = 9.81KK857 pKa = 9.97LVSLLTMVEE866 pKa = 3.92RR867 pKa = 11.84PDD869 pKa = 3.23WVRR872 pKa = 11.84QADD875 pKa = 3.58WDD877 pKa = 3.79KK878 pKa = 11.46LINKK882 pKa = 9.53RR883 pKa = 11.84LFLRR887 pKa = 11.84VNLLTDD893 pKa = 3.5TCLQEE898 pKa = 4.17YY899 pKa = 10.47CLTRR903 pKa = 11.84PSWSHH908 pKa = 6.73QYY910 pKa = 8.45TQDD913 pKa = 2.66QWEE916 pKa = 4.98DD917 pKa = 3.6YY918 pKa = 10.17LQKK921 pKa = 10.7EE922 pKa = 4.45SQAQQLVSLKK932 pKa = 8.93TQLEE936 pKa = 4.59KK937 pKa = 10.49IQQQADD943 pKa = 3.62SHH945 pKa = 6.56KK946 pKa = 10.6GSARR950 pKa = 11.84QPQAQKK956 pKa = 10.59QKK958 pKa = 10.69KK959 pKa = 9.7KK960 pKa = 10.35KK961 pKa = 9.88GKK963 pKa = 8.66QANATPVAEE972 pKa = 4.3PTTSNN977 pKa = 3.29
MM1 pKa = 7.7RR2 pKa = 11.84LCVSPFMVSCAIIIAVYY19 pKa = 6.83THH21 pKa = 6.61IGIMIGVMVGDD32 pKa = 4.15FTDD35 pKa = 3.83LPFRR39 pKa = 11.84EE40 pKa = 4.52LNKK43 pKa = 10.13QLLRR47 pKa = 11.84THH49 pKa = 6.01YY50 pKa = 10.6VHH52 pKa = 7.39AFWASLNFLGEE63 pKa = 4.53FLRR66 pKa = 11.84SMHH69 pKa = 5.89TFLWALVEE77 pKa = 5.94FILAQNFSLEE87 pKa = 4.37SVGFIALLVVPTIHH101 pKa = 5.7VLYY104 pKa = 10.84VIFRR108 pKa = 11.84PKK110 pKa = 10.09PIRR113 pKa = 11.84FGEE116 pKa = 4.01IEE118 pKa = 4.63EE119 pKa = 4.37ILDD122 pKa = 3.98GYY124 pKa = 11.0LAFTGEE130 pKa = 4.42SLQEE134 pKa = 4.0GSPLQQVQPEE144 pKa = 4.44SVPKK148 pKa = 10.14GQAVLIAEE156 pKa = 4.52TATARR161 pKa = 11.84YY162 pKa = 8.55LQGQVTRR169 pKa = 11.84MDD171 pKa = 3.59NVEE174 pKa = 4.0VTDD177 pKa = 3.87TVIEE181 pKa = 4.14RR182 pKa = 11.84WNKK185 pKa = 10.45LGIDD189 pKa = 3.66YY190 pKa = 10.72DD191 pKa = 4.2FSIVNSGEE199 pKa = 4.26TKK201 pKa = 9.11FACYY205 pKa = 9.97AGLSHH210 pKa = 6.74CTTRR214 pKa = 11.84SGKK217 pKa = 9.93YY218 pKa = 10.04FIAKK222 pKa = 10.29GNDD225 pKa = 3.15LNVQVPCYY233 pKa = 10.03SVPSLAAEE241 pKa = 3.96DD242 pKa = 4.4LMFFLVPANKK252 pKa = 9.76VSVLGNPVVKK262 pKa = 9.93PHH264 pKa = 6.15STDD267 pKa = 2.85LPANVRR273 pKa = 11.84VITSAAKK280 pKa = 9.4YY281 pKa = 10.05GEE283 pKa = 4.5KK284 pKa = 10.19FGSWSTFGRR293 pKa = 11.84LDD295 pKa = 3.68RR296 pKa = 11.84DD297 pKa = 4.12DD298 pKa = 3.68VTGCCRR304 pKa = 11.84FTGSTISGYY313 pKa = 10.78SGAGYY318 pKa = 9.49YY319 pKa = 10.57ASNAWVGIHH328 pKa = 6.53RR329 pKa = 11.84GHH331 pKa = 7.37RR332 pKa = 11.84SGLNFGFLAAYY343 pKa = 9.38VKK345 pKa = 9.27TVKK348 pKa = 10.55NVTFLDD354 pKa = 3.71KK355 pKa = 11.01AVTGEE360 pKa = 4.17SDD362 pKa = 3.43TEE364 pKa = 4.05LARR367 pKa = 11.84LLGLLRR373 pKa = 11.84EE374 pKa = 5.04DD375 pKa = 4.55GPMAYY380 pKa = 10.12SKK382 pKa = 10.77LSKK385 pKa = 10.15RR386 pKa = 11.84KK387 pKa = 10.27SGDD390 pKa = 3.24PSIDD394 pKa = 3.2LYY396 pKa = 11.03EE397 pKa = 4.05YY398 pKa = 10.74NGDD401 pKa = 3.47YY402 pKa = 10.63FYY404 pKa = 11.76VDD406 pKa = 3.36RR407 pKa = 11.84DD408 pKa = 3.99DD409 pKa = 3.94VNSYY413 pKa = 8.44QRR415 pKa = 11.84EE416 pKa = 3.75EE417 pKa = 3.85WQNYY421 pKa = 7.89LDD423 pKa = 4.87AMQMDD428 pKa = 4.8NEE430 pKa = 4.44TEE432 pKa = 3.84DD433 pKa = 3.49RR434 pKa = 11.84QVRR437 pKa = 11.84RR438 pKa = 11.84RR439 pKa = 11.84NKK441 pKa = 9.53SRR443 pKa = 11.84RR444 pKa = 11.84RR445 pKa = 11.84DD446 pKa = 3.49DD447 pKa = 3.41TDD449 pKa = 3.76DD450 pKa = 4.17FEE452 pKa = 7.14GEE454 pKa = 4.09AGSVDD459 pKa = 3.87LTHH462 pKa = 6.84LNSRR466 pKa = 11.84GGAVRR471 pKa = 11.84TEE473 pKa = 4.28CGEE476 pKa = 4.2EE477 pKa = 3.76IQGTMLLPPPRR488 pKa = 11.84CDD490 pKa = 2.86STGNVTDD497 pKa = 3.57HH498 pKa = 6.71ARR500 pKa = 11.84EE501 pKa = 3.8KK502 pKa = 10.72RR503 pKa = 11.84ADD505 pKa = 4.17EE506 pKa = 4.1IALLLIKK513 pKa = 10.53QKK515 pKa = 10.83LDD517 pKa = 3.34RR518 pKa = 11.84LSEE521 pKa = 4.01KK522 pKa = 9.91MKK524 pKa = 10.74QSFVNYY530 pKa = 9.11TFEE533 pKa = 5.17QKK535 pKa = 10.49LGWMKK540 pKa = 10.55SPQEE544 pKa = 3.84LWHH547 pKa = 6.27LTGKK551 pKa = 8.82MLSIQSALTHH561 pKa = 6.5WIKK564 pKa = 10.56PVAEE568 pKa = 4.85AYY570 pKa = 10.28SEE572 pKa = 4.25TMSRR576 pKa = 11.84VAEE579 pKa = 3.91LTPWEE584 pKa = 4.13EE585 pKa = 3.8EE586 pKa = 4.13AKK588 pKa = 10.63KK589 pKa = 10.67KK590 pKa = 9.58LAKK593 pKa = 10.25SRR595 pKa = 11.84ASYY598 pKa = 10.94NKK600 pKa = 10.53ANGTPAPPASFDD612 pKa = 4.61PILEE616 pKa = 4.08SWYY619 pKa = 9.83EE620 pKa = 3.9ATEE623 pKa = 4.01KK624 pKa = 10.61LHH626 pKa = 5.13EE627 pKa = 4.47QIMEE631 pKa = 3.69ISEE634 pKa = 4.58RR635 pKa = 11.84YY636 pKa = 8.24PPPSVTTEE644 pKa = 3.9GDD646 pKa = 3.44VADD649 pKa = 4.69PEE651 pKa = 4.54DD652 pKa = 4.39FEE654 pKa = 6.48GEE656 pKa = 4.33TGDD659 pKa = 4.46HH660 pKa = 5.69QLEE663 pKa = 4.38DD664 pKa = 3.65LLEE667 pKa = 4.18ILRR670 pKa = 11.84NPTQPPLTQPLEE682 pKa = 4.17KK683 pKa = 10.1EE684 pKa = 3.95QTSRR688 pKa = 11.84FQMKK692 pKa = 9.28HH693 pKa = 5.61LSGAPIVKK701 pKa = 8.21QTAAKK706 pKa = 10.44SPTTLPVVPAVISSGDD722 pKa = 3.79DD723 pKa = 3.5LEE725 pKa = 5.08VGADD729 pKa = 3.48AEE731 pKa = 4.63SFSASQAVSHH741 pKa = 6.09PTEE744 pKa = 4.34DD745 pKa = 3.93FIEE748 pKa = 4.72RR749 pKa = 11.84SSLTSQATSRR759 pKa = 11.84PPVLSDD765 pKa = 3.31RR766 pKa = 11.84GQRR769 pKa = 11.84HH770 pKa = 5.82ASASNQLEE778 pKa = 4.3TPLASNGQIQNDD790 pKa = 3.82QPTQLPKK797 pKa = 10.68ASAIIPEE804 pKa = 4.69CSSEE808 pKa = 4.25TTSQMSGPHH817 pKa = 5.81GMNLTKK823 pKa = 10.37SAKK826 pKa = 9.14TSGKK830 pKa = 10.23SSLSSPRR837 pKa = 11.84QEE839 pKa = 4.44SRR841 pKa = 11.84FEE843 pKa = 4.29TVWPEE848 pKa = 3.76KK849 pKa = 10.95SKK851 pKa = 9.82LTSNKK856 pKa = 9.81KK857 pKa = 9.97LVSLLTMVEE866 pKa = 3.92RR867 pKa = 11.84PDD869 pKa = 3.23WVRR872 pKa = 11.84QADD875 pKa = 3.58WDD877 pKa = 3.79KK878 pKa = 11.46LINKK882 pKa = 9.53RR883 pKa = 11.84LFLRR887 pKa = 11.84VNLLTDD893 pKa = 3.5TCLQEE898 pKa = 4.17YY899 pKa = 10.47CLTRR903 pKa = 11.84PSWSHH908 pKa = 6.73QYY910 pKa = 8.45TQDD913 pKa = 2.66QWEE916 pKa = 4.98DD917 pKa = 3.6YY918 pKa = 10.17LQKK921 pKa = 10.7EE922 pKa = 4.45SQAQQLVSLKK932 pKa = 8.93TQLEE936 pKa = 4.59KK937 pKa = 10.49IQQQADD943 pKa = 3.62SHH945 pKa = 6.56KK946 pKa = 10.6GSARR950 pKa = 11.84QPQAQKK956 pKa = 10.59QKK958 pKa = 10.69KK959 pKa = 9.7KK960 pKa = 10.35KK961 pKa = 9.88GKK963 pKa = 8.66QANATPVAEE972 pKa = 4.3PTTSNN977 pKa = 3.29
Molecular weight: 109.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE94|A0A1L3KE94_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 21 OX=1922693 PE=4 SV=1
MM1 pKa = 8.09DD2 pKa = 4.35EE3 pKa = 4.19VTSRR7 pKa = 11.84TLAPHH12 pKa = 6.59RR13 pKa = 11.84KK14 pKa = 9.12DD15 pKa = 3.09VEE17 pKa = 4.18YY18 pKa = 9.88PKK20 pKa = 10.87RR21 pKa = 11.84LDD23 pKa = 3.91TLDD26 pKa = 3.56QACSRR31 pKa = 11.84GLLRR35 pKa = 11.84DD36 pKa = 3.64NVEE39 pKa = 4.12SGGTHH44 pKa = 4.22TMGRR48 pKa = 11.84RR49 pKa = 11.84SEE51 pKa = 4.17EE52 pKa = 3.91EE53 pKa = 3.55TGKK56 pKa = 10.1IARR59 pKa = 11.84KK60 pKa = 8.58LQQSEE65 pKa = 4.61RR66 pKa = 11.84NASTSCVIRR75 pKa = 11.84PNSGVLVRR83 pKa = 11.84GNGKK87 pKa = 7.09TARR90 pKa = 11.84TDD92 pKa = 3.29NGDD95 pKa = 3.23QRR97 pKa = 11.84TLSATICDD105 pKa = 3.54NRR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84CGPGRR115 pKa = 11.84LRR117 pKa = 11.84RR118 pKa = 11.84GNGRR122 pKa = 11.84SSARR126 pKa = 11.84RR127 pKa = 11.84PVRR130 pKa = 11.84NSEE133 pKa = 4.07KK134 pKa = 10.44PNPTTVDD141 pKa = 3.25STIGEE146 pKa = 4.29GADD149 pKa = 3.11IEE151 pKa = 4.52ISNEE155 pKa = 3.73APFRR159 pKa = 11.84CPDD162 pKa = 3.62CEE164 pKa = 4.05TDD166 pKa = 3.3CRR168 pKa = 11.84KK169 pKa = 8.75ITDD172 pKa = 3.54YY173 pKa = 7.48TTRR176 pKa = 11.84CPSCDD181 pKa = 3.54KK182 pKa = 10.15QWRR185 pKa = 11.84RR186 pKa = 11.84LGGRR190 pKa = 11.84CGCRR194 pKa = 11.84EE195 pKa = 4.01LFGVPGCLTSYY206 pKa = 10.51RR207 pKa = 11.84GFHH210 pKa = 6.41RR211 pKa = 11.84KK212 pKa = 8.79IKK214 pKa = 10.36FDD216 pKa = 3.67FSSYY220 pKa = 10.64VPATCPLRR228 pKa = 11.84QGAEE232 pKa = 3.73TCEE235 pKa = 4.25CFKK238 pKa = 11.23SIGDD242 pKa = 3.6PAGFQWPDD250 pKa = 3.3PEE252 pKa = 5.61RR253 pKa = 11.84PTNPIAEE260 pKa = 4.72SICYY264 pKa = 9.87HH265 pKa = 6.11SGVFLRR271 pKa = 11.84NYY273 pKa = 9.86KK274 pKa = 10.09PDD276 pKa = 3.3VRR278 pKa = 11.84PTRR281 pKa = 11.84DD282 pKa = 3.69EE283 pKa = 4.23LDD285 pKa = 3.71QVCKK289 pKa = 10.73DD290 pKa = 3.18LWKK293 pKa = 10.42KK294 pKa = 10.41FSIFASPRR302 pKa = 11.84KK303 pKa = 9.1PVRR306 pKa = 11.84DD307 pKa = 3.54CLAGKK312 pKa = 8.76EE313 pKa = 3.98QTYY316 pKa = 9.94LQQEE320 pKa = 4.33VSKK323 pKa = 10.62PLDD326 pKa = 4.16DD327 pKa = 4.69GGKK330 pKa = 7.43TRR332 pKa = 11.84LGATSRR338 pKa = 11.84LGQVDD343 pKa = 3.7QQTSFPEE350 pKa = 4.05SEE352 pKa = 4.44PLDD355 pKa = 4.19GYY357 pKa = 11.03LPTGILPDD365 pKa = 3.52QTEE368 pKa = 4.75LEE370 pKa = 4.19SSIYY374 pKa = 9.1SRR376 pKa = 11.84PMGGLSSEE384 pKa = 4.53GEE386 pKa = 4.33SGSTTGEE393 pKa = 4.23LEE395 pKa = 3.96NSTGEE400 pKa = 4.05NPTTGGFPQGICSSTPGSKK419 pKa = 10.39AEE421 pKa = 4.14EE422 pKa = 4.44KK423 pKa = 10.31EE424 pKa = 4.07RR425 pKa = 11.84EE426 pKa = 4.13AGKK429 pKa = 10.4CYY431 pKa = 9.88PGCRR435 pKa = 11.84TDD437 pKa = 4.31HH438 pKa = 5.87EE439 pKa = 4.36QLVRR443 pKa = 11.84GILPSFDD450 pKa = 5.18DD451 pKa = 4.01FTTLVNRR458 pKa = 11.84QLDD461 pKa = 3.93KK462 pKa = 11.46NSTPGIPYY470 pKa = 8.45NTIAPTNAEE479 pKa = 3.93VFGWDD484 pKa = 3.23GYY486 pKa = 11.65KK487 pKa = 8.9YY488 pKa = 9.35TKK490 pKa = 10.45NLDD493 pKa = 3.25MLYY496 pKa = 10.4NDD498 pKa = 3.31VFEE501 pKa = 5.57RR502 pKa = 11.84MVDD505 pKa = 3.91LLSGPASNPINLFLKK520 pKa = 10.32PEE522 pKa = 3.83PHH524 pKa = 6.83KK525 pKa = 10.76KK526 pKa = 9.99SKK528 pKa = 10.94CEE530 pKa = 3.57AGAWRR535 pKa = 11.84IIWGCSCVDD544 pKa = 3.27QMVALVLWYY553 pKa = 9.85PNVDD557 pKa = 3.51KK558 pKa = 10.91IVNQWGMSPSMIGWTPQRR576 pKa = 11.84GGHH579 pKa = 5.7KK580 pKa = 9.52WLSEE584 pKa = 4.06LFNARR589 pKa = 11.84HH590 pKa = 6.06EE591 pKa = 4.55SVQCADD597 pKa = 3.11KK598 pKa = 11.18SAWDD602 pKa = 3.33WTVQFWVCVAVSYY615 pKa = 10.88VLVNLMVGASEE626 pKa = 4.04AQRR629 pKa = 11.84VIAHH633 pKa = 5.57NHH635 pKa = 4.66CMAMLGYY642 pKa = 8.45QTVQFGKK649 pKa = 10.53KK650 pKa = 9.04KK651 pKa = 10.74LKK653 pKa = 9.96FQIPGRR659 pKa = 11.84LPSGWFWTIFFNCAGQASVDD679 pKa = 3.7SLASQRR685 pKa = 11.84VGEE688 pKa = 4.31VPDD691 pKa = 4.47PKK693 pKa = 10.48LKK695 pKa = 11.17AMGDD699 pKa = 3.59DD700 pKa = 3.47TAQTVRR706 pKa = 11.84SDD708 pKa = 4.17AYY710 pKa = 9.1WKK712 pKa = 10.54AVNQLCIVKK721 pKa = 10.48QIDD724 pKa = 3.52TFKK727 pKa = 10.93PGDD730 pKa = 3.55EE731 pKa = 4.69FEE733 pKa = 4.55FCGTLMNHH741 pKa = 6.09EE742 pKa = 4.88RR743 pKa = 11.84GTPAKK748 pKa = 9.75RR749 pKa = 11.84GSHH752 pKa = 6.45AFRR755 pKa = 11.84LHH757 pKa = 6.76FLRR760 pKa = 11.84DD761 pKa = 3.08EE762 pKa = 4.45CAAEE766 pKa = 4.12GLVSYY771 pKa = 10.98ALLYY775 pKa = 10.87AHH777 pKa = 7.48DD778 pKa = 4.32PAAFATISALQCEE791 pKa = 4.61IGRR794 pKa = 11.84YY795 pKa = 7.35DD796 pKa = 3.55TLITRR801 pKa = 11.84DD802 pKa = 3.86AAVAWYY808 pKa = 10.19SGTALGIGG816 pKa = 3.61
MM1 pKa = 8.09DD2 pKa = 4.35EE3 pKa = 4.19VTSRR7 pKa = 11.84TLAPHH12 pKa = 6.59RR13 pKa = 11.84KK14 pKa = 9.12DD15 pKa = 3.09VEE17 pKa = 4.18YY18 pKa = 9.88PKK20 pKa = 10.87RR21 pKa = 11.84LDD23 pKa = 3.91TLDD26 pKa = 3.56QACSRR31 pKa = 11.84GLLRR35 pKa = 11.84DD36 pKa = 3.64NVEE39 pKa = 4.12SGGTHH44 pKa = 4.22TMGRR48 pKa = 11.84RR49 pKa = 11.84SEE51 pKa = 4.17EE52 pKa = 3.91EE53 pKa = 3.55TGKK56 pKa = 10.1IARR59 pKa = 11.84KK60 pKa = 8.58LQQSEE65 pKa = 4.61RR66 pKa = 11.84NASTSCVIRR75 pKa = 11.84PNSGVLVRR83 pKa = 11.84GNGKK87 pKa = 7.09TARR90 pKa = 11.84TDD92 pKa = 3.29NGDD95 pKa = 3.23QRR97 pKa = 11.84TLSATICDD105 pKa = 3.54NRR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84CGPGRR115 pKa = 11.84LRR117 pKa = 11.84RR118 pKa = 11.84GNGRR122 pKa = 11.84SSARR126 pKa = 11.84RR127 pKa = 11.84PVRR130 pKa = 11.84NSEE133 pKa = 4.07KK134 pKa = 10.44PNPTTVDD141 pKa = 3.25STIGEE146 pKa = 4.29GADD149 pKa = 3.11IEE151 pKa = 4.52ISNEE155 pKa = 3.73APFRR159 pKa = 11.84CPDD162 pKa = 3.62CEE164 pKa = 4.05TDD166 pKa = 3.3CRR168 pKa = 11.84KK169 pKa = 8.75ITDD172 pKa = 3.54YY173 pKa = 7.48TTRR176 pKa = 11.84CPSCDD181 pKa = 3.54KK182 pKa = 10.15QWRR185 pKa = 11.84RR186 pKa = 11.84LGGRR190 pKa = 11.84CGCRR194 pKa = 11.84EE195 pKa = 4.01LFGVPGCLTSYY206 pKa = 10.51RR207 pKa = 11.84GFHH210 pKa = 6.41RR211 pKa = 11.84KK212 pKa = 8.79IKK214 pKa = 10.36FDD216 pKa = 3.67FSSYY220 pKa = 10.64VPATCPLRR228 pKa = 11.84QGAEE232 pKa = 3.73TCEE235 pKa = 4.25CFKK238 pKa = 11.23SIGDD242 pKa = 3.6PAGFQWPDD250 pKa = 3.3PEE252 pKa = 5.61RR253 pKa = 11.84PTNPIAEE260 pKa = 4.72SICYY264 pKa = 9.87HH265 pKa = 6.11SGVFLRR271 pKa = 11.84NYY273 pKa = 9.86KK274 pKa = 10.09PDD276 pKa = 3.3VRR278 pKa = 11.84PTRR281 pKa = 11.84DD282 pKa = 3.69EE283 pKa = 4.23LDD285 pKa = 3.71QVCKK289 pKa = 10.73DD290 pKa = 3.18LWKK293 pKa = 10.42KK294 pKa = 10.41FSIFASPRR302 pKa = 11.84KK303 pKa = 9.1PVRR306 pKa = 11.84DD307 pKa = 3.54CLAGKK312 pKa = 8.76EE313 pKa = 3.98QTYY316 pKa = 9.94LQQEE320 pKa = 4.33VSKK323 pKa = 10.62PLDD326 pKa = 4.16DD327 pKa = 4.69GGKK330 pKa = 7.43TRR332 pKa = 11.84LGATSRR338 pKa = 11.84LGQVDD343 pKa = 3.7QQTSFPEE350 pKa = 4.05SEE352 pKa = 4.44PLDD355 pKa = 4.19GYY357 pKa = 11.03LPTGILPDD365 pKa = 3.52QTEE368 pKa = 4.75LEE370 pKa = 4.19SSIYY374 pKa = 9.1SRR376 pKa = 11.84PMGGLSSEE384 pKa = 4.53GEE386 pKa = 4.33SGSTTGEE393 pKa = 4.23LEE395 pKa = 3.96NSTGEE400 pKa = 4.05NPTTGGFPQGICSSTPGSKK419 pKa = 10.39AEE421 pKa = 4.14EE422 pKa = 4.44KK423 pKa = 10.31EE424 pKa = 4.07RR425 pKa = 11.84EE426 pKa = 4.13AGKK429 pKa = 10.4CYY431 pKa = 9.88PGCRR435 pKa = 11.84TDD437 pKa = 4.31HH438 pKa = 5.87EE439 pKa = 4.36QLVRR443 pKa = 11.84GILPSFDD450 pKa = 5.18DD451 pKa = 4.01FTTLVNRR458 pKa = 11.84QLDD461 pKa = 3.93KK462 pKa = 11.46NSTPGIPYY470 pKa = 8.45NTIAPTNAEE479 pKa = 3.93VFGWDD484 pKa = 3.23GYY486 pKa = 11.65KK487 pKa = 8.9YY488 pKa = 9.35TKK490 pKa = 10.45NLDD493 pKa = 3.25MLYY496 pKa = 10.4NDD498 pKa = 3.31VFEE501 pKa = 5.57RR502 pKa = 11.84MVDD505 pKa = 3.91LLSGPASNPINLFLKK520 pKa = 10.32PEE522 pKa = 3.83PHH524 pKa = 6.83KK525 pKa = 10.76KK526 pKa = 9.99SKK528 pKa = 10.94CEE530 pKa = 3.57AGAWRR535 pKa = 11.84IIWGCSCVDD544 pKa = 3.27QMVALVLWYY553 pKa = 9.85PNVDD557 pKa = 3.51KK558 pKa = 10.91IVNQWGMSPSMIGWTPQRR576 pKa = 11.84GGHH579 pKa = 5.7KK580 pKa = 9.52WLSEE584 pKa = 4.06LFNARR589 pKa = 11.84HH590 pKa = 6.06EE591 pKa = 4.55SVQCADD597 pKa = 3.11KK598 pKa = 11.18SAWDD602 pKa = 3.33WTVQFWVCVAVSYY615 pKa = 10.88VLVNLMVGASEE626 pKa = 4.04AQRR629 pKa = 11.84VIAHH633 pKa = 5.57NHH635 pKa = 4.66CMAMLGYY642 pKa = 8.45QTVQFGKK649 pKa = 10.53KK650 pKa = 9.04KK651 pKa = 10.74LKK653 pKa = 9.96FQIPGRR659 pKa = 11.84LPSGWFWTIFFNCAGQASVDD679 pKa = 3.7SLASQRR685 pKa = 11.84VGEE688 pKa = 4.31VPDD691 pKa = 4.47PKK693 pKa = 10.48LKK695 pKa = 11.17AMGDD699 pKa = 3.59DD700 pKa = 3.47TAQTVRR706 pKa = 11.84SDD708 pKa = 4.17AYY710 pKa = 9.1WKK712 pKa = 10.54AVNQLCIVKK721 pKa = 10.48QIDD724 pKa = 3.52TFKK727 pKa = 10.93PGDD730 pKa = 3.55EE731 pKa = 4.69FEE733 pKa = 4.55FCGTLMNHH741 pKa = 6.09EE742 pKa = 4.88RR743 pKa = 11.84GTPAKK748 pKa = 9.75RR749 pKa = 11.84GSHH752 pKa = 6.45AFRR755 pKa = 11.84LHH757 pKa = 6.76FLRR760 pKa = 11.84DD761 pKa = 3.08EE762 pKa = 4.45CAAEE766 pKa = 4.12GLVSYY771 pKa = 10.98ALLYY775 pKa = 10.87AHH777 pKa = 7.48DD778 pKa = 4.32PAAFATISALQCEE791 pKa = 4.61IGRR794 pKa = 11.84YY795 pKa = 7.35DD796 pKa = 3.55TLITRR801 pKa = 11.84DD802 pKa = 3.86AAVAWYY808 pKa = 10.19SGTALGIGG816 pKa = 3.61
Molecular weight: 90.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1793 |
816 |
977 |
896.5 |
100.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.86 ± 0.244 | 2.454 ± 0.981 |
5.745 ± 0.256 | 6.581 ± 0.467 |
3.625 ± 0.034 | 7.195 ± 1.089 |
1.952 ± 0.158 | 3.96 ± 0.026 |
5.521 ± 0.25 | 8.254 ± 0.767 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.84 ± 0.165 | 3.569 ± 0.072 |
5.745 ± 0.256 | 4.908 ± 0.578 |
6.247 ± 0.822 | 8.199 ± 0.565 |
6.804 ± 0.043 | 5.856 ± 0.31 |
1.84 ± 0.162 | 2.844 ± 0.099 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
