
Thermoplasma acidophilum (strain ATCC 25905 / DSM 1728 / JCM 9062 / NBRC 15155 / AMRC-C165)
Taxonomy: cellular organisms; Archaea; Candidatus Thermoplasmatota; Thermoplasmata; Thermoplasmatales; Thermoplasmataceae; Thermoplasma; Thermoplasma acidophilum
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
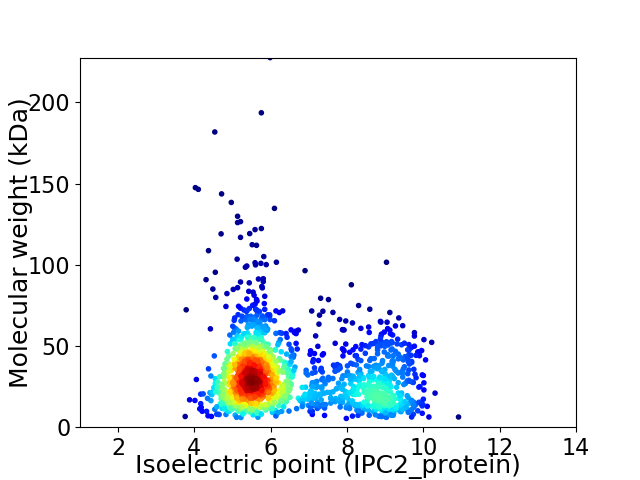
Virtual 2D-PAGE plot for 1482 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9HIP0|RL37A_THEAC 50S ribosomal protein L37Ae OS=Thermoplasma acidophilum (strain ATCC 25905 / DSM 1728 / JCM 9062 / NBRC 15155 / AMRC-C165) OX=273075 GN=rpl37ae PE=3 SV=1
MM1 pKa = 6.7NTEE4 pKa = 3.86HH5 pKa = 6.13TVSVFILIFLFIGSYY20 pKa = 9.56SAPGSISTIHH30 pKa = 5.82QPAYY34 pKa = 9.24VQNTKK39 pKa = 9.62MSANNTSEE47 pKa = 4.21TIDD50 pKa = 4.78LINDD54 pKa = 3.62SVISGNFMDD63 pKa = 4.45TGNAIVPTSGAYY75 pKa = 10.02ASDD78 pKa = 3.62CGCYY82 pKa = 9.53FVANYY87 pKa = 7.49FTDD90 pKa = 3.24NVYY93 pKa = 10.9VINATTYY100 pKa = 10.26RR101 pKa = 11.84IANVMPVGSGPISVMYY117 pKa = 10.61VEE119 pKa = 5.26SNGLVYY125 pKa = 10.49VANQNSGDD133 pKa = 3.22ISVINTHH140 pKa = 6.13LMSVIDD146 pKa = 4.33TIGVGSGPQSIAYY159 pKa = 9.07DD160 pKa = 3.74PQIDD164 pKa = 3.83EE165 pKa = 4.43VFIANSYY172 pKa = 8.98SGSVTVISGSNSVVANISVAGASGVAYY199 pKa = 9.98SQYY202 pKa = 10.91NGYY205 pKa = 10.05IYY207 pKa = 10.54VSQYY211 pKa = 8.14GHH213 pKa = 6.16GMVSVINPLTLSTVGTFEE231 pKa = 4.94AGSSPSSLAIDD242 pKa = 3.68NLNGKK247 pKa = 8.22VFIDD251 pKa = 3.7NQGSDD256 pKa = 4.27TITVTGPNGTYY267 pKa = 9.94IRR269 pKa = 11.84TIPVGSDD276 pKa = 2.9PLQVMFCNYY285 pKa = 9.62SDD287 pKa = 3.8QVYY290 pKa = 8.85TSNFDD295 pKa = 3.45SDD297 pKa = 3.92TVTAINAINFSSITNISVGLNPYY320 pKa = 10.0GLFMGNGRR328 pKa = 11.84NEE330 pKa = 3.48IAVANEE336 pKa = 3.62YY337 pKa = 9.89TNNISFINTSTDD349 pKa = 2.81EE350 pKa = 4.29VYY352 pKa = 11.11YY353 pKa = 10.85SLTIGSGPQGLAYY366 pKa = 10.48DD367 pKa = 4.21PVNSYY372 pKa = 11.3LYY374 pKa = 9.0VTNAYY379 pKa = 10.87SNFVYY384 pKa = 10.78VLDD387 pKa = 4.23KK388 pKa = 10.87NYY390 pKa = 11.04SVIAKK395 pKa = 9.49VYY397 pKa = 9.72VGWGPSGIAYY407 pKa = 9.78DD408 pKa = 3.98PYY410 pKa = 11.1NNYY413 pKa = 10.05VYY415 pKa = 10.57VANYY419 pKa = 7.82DD420 pKa = 3.56TDD422 pKa = 3.78NVSVLDD428 pKa = 4.0TQTQTVLRR436 pKa = 11.84NIAVGEE442 pKa = 4.5GPAGIVVNPSNGYY455 pKa = 9.58VYY457 pKa = 10.56SINQLSDD464 pKa = 3.51DD465 pKa = 3.81VSVINPSNNSVISSIITGSSPFNGVYY491 pKa = 10.33DD492 pKa = 3.67QANGFIYY499 pKa = 10.65VGNFGSNNISVINPDD514 pKa = 3.64SEE516 pKa = 4.66SVVYY520 pKa = 8.79TIQVGSGPQDD530 pKa = 3.36LSYY533 pKa = 11.2DD534 pKa = 3.67PEE536 pKa = 4.84NGYY539 pKa = 10.3IYY541 pKa = 10.37VINFNSDD548 pKa = 2.88SVSIINGKK556 pKa = 8.65EE557 pKa = 3.79RR558 pKa = 11.84VLSTINTGSGPEE570 pKa = 4.36GICYY574 pKa = 10.24DD575 pKa = 3.95PYY577 pKa = 11.72DD578 pKa = 3.7NSLYY582 pKa = 10.67VSNSGSNDD590 pKa = 2.97VYY592 pKa = 11.23VINATTEE599 pKa = 4.02GVEE602 pKa = 4.21AIIPVGIYY610 pKa = 9.46PFRR613 pKa = 11.84DD614 pKa = 3.42CVDD617 pKa = 3.67PANGLVFVANTNSGTVSIIEE637 pKa = 4.22PMSPTAGTVISPHH650 pKa = 5.05VQNDD654 pKa = 3.22ALFFILAILVCALAITGYY672 pKa = 10.21IATIKK677 pKa = 10.09HH678 pKa = 5.56KK679 pKa = 10.64KK680 pKa = 8.65
MM1 pKa = 6.7NTEE4 pKa = 3.86HH5 pKa = 6.13TVSVFILIFLFIGSYY20 pKa = 9.56SAPGSISTIHH30 pKa = 5.82QPAYY34 pKa = 9.24VQNTKK39 pKa = 9.62MSANNTSEE47 pKa = 4.21TIDD50 pKa = 4.78LINDD54 pKa = 3.62SVISGNFMDD63 pKa = 4.45TGNAIVPTSGAYY75 pKa = 10.02ASDD78 pKa = 3.62CGCYY82 pKa = 9.53FVANYY87 pKa = 7.49FTDD90 pKa = 3.24NVYY93 pKa = 10.9VINATTYY100 pKa = 10.26RR101 pKa = 11.84IANVMPVGSGPISVMYY117 pKa = 10.61VEE119 pKa = 5.26SNGLVYY125 pKa = 10.49VANQNSGDD133 pKa = 3.22ISVINTHH140 pKa = 6.13LMSVIDD146 pKa = 4.33TIGVGSGPQSIAYY159 pKa = 9.07DD160 pKa = 3.74PQIDD164 pKa = 3.83EE165 pKa = 4.43VFIANSYY172 pKa = 8.98SGSVTVISGSNSVVANISVAGASGVAYY199 pKa = 9.98SQYY202 pKa = 10.91NGYY205 pKa = 10.05IYY207 pKa = 10.54VSQYY211 pKa = 8.14GHH213 pKa = 6.16GMVSVINPLTLSTVGTFEE231 pKa = 4.94AGSSPSSLAIDD242 pKa = 3.68NLNGKK247 pKa = 8.22VFIDD251 pKa = 3.7NQGSDD256 pKa = 4.27TITVTGPNGTYY267 pKa = 9.94IRR269 pKa = 11.84TIPVGSDD276 pKa = 2.9PLQVMFCNYY285 pKa = 9.62SDD287 pKa = 3.8QVYY290 pKa = 8.85TSNFDD295 pKa = 3.45SDD297 pKa = 3.92TVTAINAINFSSITNISVGLNPYY320 pKa = 10.0GLFMGNGRR328 pKa = 11.84NEE330 pKa = 3.48IAVANEE336 pKa = 3.62YY337 pKa = 9.89TNNISFINTSTDD349 pKa = 2.81EE350 pKa = 4.29VYY352 pKa = 11.11YY353 pKa = 10.85SLTIGSGPQGLAYY366 pKa = 10.48DD367 pKa = 4.21PVNSYY372 pKa = 11.3LYY374 pKa = 9.0VTNAYY379 pKa = 10.87SNFVYY384 pKa = 10.78VLDD387 pKa = 4.23KK388 pKa = 10.87NYY390 pKa = 11.04SVIAKK395 pKa = 9.49VYY397 pKa = 9.72VGWGPSGIAYY407 pKa = 9.78DD408 pKa = 3.98PYY410 pKa = 11.1NNYY413 pKa = 10.05VYY415 pKa = 10.57VANYY419 pKa = 7.82DD420 pKa = 3.56TDD422 pKa = 3.78NVSVLDD428 pKa = 4.0TQTQTVLRR436 pKa = 11.84NIAVGEE442 pKa = 4.5GPAGIVVNPSNGYY455 pKa = 9.58VYY457 pKa = 10.56SINQLSDD464 pKa = 3.51DD465 pKa = 3.81VSVINPSNNSVISSIITGSSPFNGVYY491 pKa = 10.33DD492 pKa = 3.67QANGFIYY499 pKa = 10.65VGNFGSNNISVINPDD514 pKa = 3.64SEE516 pKa = 4.66SVVYY520 pKa = 8.79TIQVGSGPQDD530 pKa = 3.36LSYY533 pKa = 11.2DD534 pKa = 3.67PEE536 pKa = 4.84NGYY539 pKa = 10.3IYY541 pKa = 10.37VINFNSDD548 pKa = 2.88SVSIINGKK556 pKa = 8.65EE557 pKa = 3.79RR558 pKa = 11.84VLSTINTGSGPEE570 pKa = 4.36GICYY574 pKa = 10.24DD575 pKa = 3.95PYY577 pKa = 11.72DD578 pKa = 3.7NSLYY582 pKa = 10.67VSNSGSNDD590 pKa = 2.97VYY592 pKa = 11.23VINATTEE599 pKa = 4.02GVEE602 pKa = 4.21AIIPVGIYY610 pKa = 9.46PFRR613 pKa = 11.84DD614 pKa = 3.42CVDD617 pKa = 3.67PANGLVFVANTNSGTVSIIEE637 pKa = 4.22PMSPTAGTVISPHH650 pKa = 5.05VQNDD654 pKa = 3.22ALFFILAILVCALAITGYY672 pKa = 10.21IATIKK677 pKa = 10.09HH678 pKa = 5.56KK679 pKa = 10.64KK680 pKa = 8.65
Molecular weight: 72.25 kDa
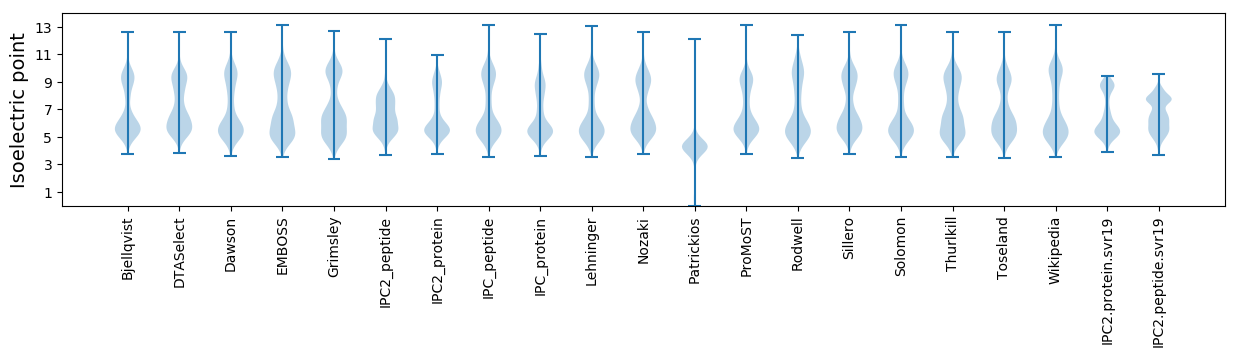
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9HM23|Q9HM23_THEAC Phosphate permease related protein OS=Thermoplasma acidophilum (strain ATCC 25905 / DSM 1728 / JCM 9062 / NBRC 15155 / AMRC-C165) OX=273075 GN=Ta0048 PE=4 SV=1
MM1 pKa = 7.4SRR3 pKa = 11.84NKK5 pKa = 10.27EE6 pKa = 3.86LGRR9 pKa = 11.84KK10 pKa = 7.91IRR12 pKa = 11.84LMKK15 pKa = 10.23KK16 pKa = 8.44IKK18 pKa = 9.27QNRR21 pKa = 11.84RR22 pKa = 11.84VPGWVMMRR30 pKa = 11.84TARR33 pKa = 11.84KK34 pKa = 7.94VTQNPLRR41 pKa = 11.84RR42 pKa = 11.84NWRR45 pKa = 11.84RR46 pKa = 11.84GSLKK50 pKa = 10.12II51 pKa = 4.26
MM1 pKa = 7.4SRR3 pKa = 11.84NKK5 pKa = 10.27EE6 pKa = 3.86LGRR9 pKa = 11.84KK10 pKa = 7.91IRR12 pKa = 11.84LMKK15 pKa = 10.23KK16 pKa = 8.44IKK18 pKa = 9.27QNRR21 pKa = 11.84RR22 pKa = 11.84VPGWVMMRR30 pKa = 11.84TARR33 pKa = 11.84KK34 pKa = 7.94VTQNPLRR41 pKa = 11.84RR42 pKa = 11.84NWRR45 pKa = 11.84RR46 pKa = 11.84GSLKK50 pKa = 10.12II51 pKa = 4.26
Molecular weight: 6.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
453232 |
45 |
2081 |
305.8 |
34.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.96 ± 0.055 | 0.601 ± 0.018 |
5.751 ± 0.061 | 5.984 ± 0.079 |
4.701 ± 0.056 | 7.251 ± 0.054 |
1.646 ± 0.02 | 9.021 ± 0.055 |
5.641 ± 0.068 | 8.384 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.195 ± 0.032 | 4.251 ± 0.053 |
3.963 ± 0.039 | 2.157 ± 0.031 |
5.499 ± 0.07 | 7.568 ± 0.061 |
4.779 ± 0.05 | 7.157 ± 0.052 |
0.854 ± 0.023 | 4.635 ± 0.053 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
