
Anaerotruncus sp. CAG:528
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Anaerotruncus; environmental samples
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
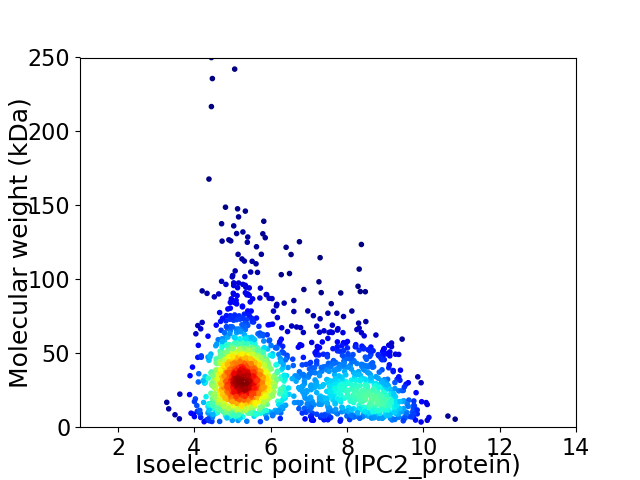
Virtual 2D-PAGE plot for 1705 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5Y696|R5Y696_9FIRM Dinucleotide-utilizing enzymes involved in molybdopterin and thiamine biosynthesis family 1 OS=Anaerotruncus sp. CAG:528 OX=1262700 GN=BN695_00471 PE=4 SV=1
MM1 pKa = 7.87ADD3 pKa = 3.67YY4 pKa = 10.98EE5 pKa = 4.51NNLNNTEE12 pKa = 4.28TTSGGAPQPNTNAQVQPASEE32 pKa = 4.29PKK34 pKa = 9.76PEE36 pKa = 4.43QINEE40 pKa = 4.11PAQGAQSAPSYY51 pKa = 10.95AEE53 pKa = 3.93PQGQSYY59 pKa = 7.22TTSPQEE65 pKa = 3.81DD66 pKa = 4.27TYY68 pKa = 11.58YY69 pKa = 11.17APQANQAPQGNYY81 pKa = 8.33NQPGYY86 pKa = 9.65QAPPQQAYY94 pKa = 8.84YY95 pKa = 10.6APQNGAQVPPYY106 pKa = 10.79GMPQQPVEE114 pKa = 4.31QKK116 pKa = 11.02ASVGLAILSFFIPIAGLIIFLTKK139 pKa = 10.63KK140 pKa = 10.15NDD142 pKa = 3.49RR143 pKa = 11.84PKK145 pKa = 8.33TAKK148 pKa = 10.37VSGICALVSFILNIVIGVIGGVIGGVAATKK178 pKa = 10.1IAEE181 pKa = 4.56NPDD184 pKa = 3.29QLDD187 pKa = 3.64SYY189 pKa = 11.11ISDD192 pKa = 4.82AIGDD196 pKa = 3.81NSDD199 pKa = 3.55NLVDD203 pKa = 4.78GDD205 pKa = 3.84EE206 pKa = 4.92LGNFVCDD213 pKa = 3.51VTDD216 pKa = 3.48VSKK219 pKa = 8.23TTDD222 pKa = 3.32ANGKK226 pKa = 7.82PAIIVTYY233 pKa = 8.62YY234 pKa = 10.49FKK236 pKa = 11.29NNSDD240 pKa = 3.43TAIDD244 pKa = 3.99FDD246 pKa = 4.09SALYY250 pKa = 9.73TIVKK254 pKa = 10.35QNDD257 pKa = 2.99NTLIRR262 pKa = 11.84TVLGTSDD269 pKa = 3.96DD270 pKa = 3.71TDD272 pKa = 4.07LADD275 pKa = 3.82TTATEE280 pKa = 4.35PGATSKK286 pKa = 10.79VKK288 pKa = 10.23RR289 pKa = 11.84AYY291 pKa = 9.08TLNDD295 pKa = 3.05QTSDD299 pKa = 3.78VVFEE303 pKa = 5.29LYY305 pKa = 10.93DD306 pKa = 3.6NTDD309 pKa = 3.75EE310 pKa = 4.41NNMYY314 pKa = 10.05YY315 pKa = 10.62LEE317 pKa = 4.2YY318 pKa = 10.66TFTLGNN324 pKa = 3.53
MM1 pKa = 7.87ADD3 pKa = 3.67YY4 pKa = 10.98EE5 pKa = 4.51NNLNNTEE12 pKa = 4.28TTSGGAPQPNTNAQVQPASEE32 pKa = 4.29PKK34 pKa = 9.76PEE36 pKa = 4.43QINEE40 pKa = 4.11PAQGAQSAPSYY51 pKa = 10.95AEE53 pKa = 3.93PQGQSYY59 pKa = 7.22TTSPQEE65 pKa = 3.81DD66 pKa = 4.27TYY68 pKa = 11.58YY69 pKa = 11.17APQANQAPQGNYY81 pKa = 8.33NQPGYY86 pKa = 9.65QAPPQQAYY94 pKa = 8.84YY95 pKa = 10.6APQNGAQVPPYY106 pKa = 10.79GMPQQPVEE114 pKa = 4.31QKK116 pKa = 11.02ASVGLAILSFFIPIAGLIIFLTKK139 pKa = 10.63KK140 pKa = 10.15NDD142 pKa = 3.49RR143 pKa = 11.84PKK145 pKa = 8.33TAKK148 pKa = 10.37VSGICALVSFILNIVIGVIGGVIGGVAATKK178 pKa = 10.1IAEE181 pKa = 4.56NPDD184 pKa = 3.29QLDD187 pKa = 3.64SYY189 pKa = 11.11ISDD192 pKa = 4.82AIGDD196 pKa = 3.81NSDD199 pKa = 3.55NLVDD203 pKa = 4.78GDD205 pKa = 3.84EE206 pKa = 4.92LGNFVCDD213 pKa = 3.51VTDD216 pKa = 3.48VSKK219 pKa = 8.23TTDD222 pKa = 3.32ANGKK226 pKa = 7.82PAIIVTYY233 pKa = 8.62YY234 pKa = 10.49FKK236 pKa = 11.29NNSDD240 pKa = 3.43TAIDD244 pKa = 3.99FDD246 pKa = 4.09SALYY250 pKa = 9.73TIVKK254 pKa = 10.35QNDD257 pKa = 2.99NTLIRR262 pKa = 11.84TVLGTSDD269 pKa = 3.96DD270 pKa = 3.71TDD272 pKa = 4.07LADD275 pKa = 3.82TTATEE280 pKa = 4.35PGATSKK286 pKa = 10.79VKK288 pKa = 10.23RR289 pKa = 11.84AYY291 pKa = 9.08TLNDD295 pKa = 3.05QTSDD299 pKa = 3.78VVFEE303 pKa = 5.29LYY305 pKa = 10.93DD306 pKa = 3.6NTDD309 pKa = 3.75EE310 pKa = 4.41NNMYY314 pKa = 10.05YY315 pKa = 10.62LEE317 pKa = 4.2YY318 pKa = 10.66TFTLGNN324 pKa = 3.53
Molecular weight: 34.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5XWP9|R5XWP9_9FIRM Predicted spore germination protein OS=Anaerotruncus sp. CAG:528 OX=1262700 GN=BN695_01562 PE=3 SV=1
MM1 pKa = 7.78IKK3 pKa = 9.22GTNRR7 pKa = 11.84KK8 pKa = 9.06CVFTPGYY15 pKa = 8.97KK16 pKa = 9.6PRR18 pKa = 11.84NIAIGVPRR26 pKa = 11.84NRR28 pKa = 11.84GRR30 pKa = 11.84GLPRR34 pKa = 11.84TVLRR38 pKa = 11.84FIPNLISVCPAYY50 pKa = 10.07GVPINMRR57 pKa = 11.84RR58 pKa = 11.84AAGLGINGKK67 pKa = 8.01ILRR70 pKa = 11.84CRR72 pKa = 11.84QRR74 pKa = 11.84IAVFNLHH81 pKa = 6.48ARR83 pKa = 11.84VFMPTAAHH91 pKa = 5.97CFVYY95 pKa = 10.68SVFNRR100 pKa = 11.84IRR102 pKa = 11.84IEE104 pKa = 3.68RR105 pKa = 11.84GRR107 pKa = 11.84ASKK110 pKa = 10.25FNKK113 pKa = 9.89RR114 pKa = 11.84GFCAQICLCPTEE126 pKa = 4.85NISLIYY132 pKa = 10.4SVMVALISGKK142 pKa = 9.97IICASVNAYY151 pKa = 10.14QIGITEE157 pKa = 4.56HH158 pKa = 6.61GIKK161 pKa = 8.9PTVKK165 pKa = 9.66INVNIHH171 pKa = 6.55FIFIHH176 pKa = 6.88KK177 pKa = 9.31IAQGAAFARR186 pKa = 11.84LEE188 pKa = 4.01FARR191 pKa = 11.84YY192 pKa = 7.44EE193 pKa = 4.28IPVHH197 pKa = 5.49IHH199 pKa = 5.96IIRR202 pKa = 11.84RR203 pKa = 11.84ASAEE207 pKa = 4.1CTGFCAVVVFAVNSAIRR224 pKa = 11.84IGAGHH229 pKa = 6.51YY230 pKa = 9.41NQTIIPKK237 pKa = 9.48IIRR240 pKa = 11.84IFGEE244 pKa = 3.59IFHH247 pKa = 6.65NVICGINKK255 pKa = 8.77QFTVSVITIFKK266 pKa = 8.18TVNRR270 pKa = 11.84KK271 pKa = 7.11GTKK274 pKa = 8.95QSFAVVAAAVYY285 pKa = 10.26NKK287 pKa = 9.82RR288 pKa = 11.84LQIAAEE294 pKa = 4.01RR295 pKa = 11.84TFAINDD301 pKa = 3.38FCYY304 pKa = 10.32II305 pKa = 3.78
MM1 pKa = 7.78IKK3 pKa = 9.22GTNRR7 pKa = 11.84KK8 pKa = 9.06CVFTPGYY15 pKa = 8.97KK16 pKa = 9.6PRR18 pKa = 11.84NIAIGVPRR26 pKa = 11.84NRR28 pKa = 11.84GRR30 pKa = 11.84GLPRR34 pKa = 11.84TVLRR38 pKa = 11.84FIPNLISVCPAYY50 pKa = 10.07GVPINMRR57 pKa = 11.84RR58 pKa = 11.84AAGLGINGKK67 pKa = 8.01ILRR70 pKa = 11.84CRR72 pKa = 11.84QRR74 pKa = 11.84IAVFNLHH81 pKa = 6.48ARR83 pKa = 11.84VFMPTAAHH91 pKa = 5.97CFVYY95 pKa = 10.68SVFNRR100 pKa = 11.84IRR102 pKa = 11.84IEE104 pKa = 3.68RR105 pKa = 11.84GRR107 pKa = 11.84ASKK110 pKa = 10.25FNKK113 pKa = 9.89RR114 pKa = 11.84GFCAQICLCPTEE126 pKa = 4.85NISLIYY132 pKa = 10.4SVMVALISGKK142 pKa = 9.97IICASVNAYY151 pKa = 10.14QIGITEE157 pKa = 4.56HH158 pKa = 6.61GIKK161 pKa = 8.9PTVKK165 pKa = 9.66INVNIHH171 pKa = 6.55FIFIHH176 pKa = 6.88KK177 pKa = 9.31IAQGAAFARR186 pKa = 11.84LEE188 pKa = 4.01FARR191 pKa = 11.84YY192 pKa = 7.44EE193 pKa = 4.28IPVHH197 pKa = 5.49IHH199 pKa = 5.96IIRR202 pKa = 11.84RR203 pKa = 11.84ASAEE207 pKa = 4.1CTGFCAVVVFAVNSAIRR224 pKa = 11.84IGAGHH229 pKa = 6.51YY230 pKa = 9.41NQTIIPKK237 pKa = 9.48IIRR240 pKa = 11.84IFGEE244 pKa = 3.59IFHH247 pKa = 6.65NVICGINKK255 pKa = 8.77QFTVSVITIFKK266 pKa = 8.18TVNRR270 pKa = 11.84KK271 pKa = 7.11GTKK274 pKa = 8.95QSFAVVAAAVYY285 pKa = 10.26NKK287 pKa = 9.82RR288 pKa = 11.84LQIAAEE294 pKa = 4.01RR295 pKa = 11.84TFAINDD301 pKa = 3.38FCYY304 pKa = 10.32II305 pKa = 3.78
Molecular weight: 34.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
549919 |
32 |
2274 |
322.5 |
35.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.707 ± 0.062 | 1.817 ± 0.025 |
5.798 ± 0.055 | 6.699 ± 0.065 |
4.431 ± 0.043 | 7.176 ± 0.049 |
1.481 ± 0.025 | 7.429 ± 0.059 |
7.592 ± 0.04 | 8.21 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.607 ± 0.026 | 5.082 ± 0.052 |
3.317 ± 0.033 | 2.597 ± 0.029 |
3.697 ± 0.046 | 6.455 ± 0.059 |
5.217 ± 0.061 | 6.896 ± 0.049 |
0.769 ± 0.021 | 4.022 ± 0.049 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
