
Deinococcus soli Cha et al. 2016
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Deinococcus-Thermus; Deinococci; Deinococcales; Deinococcaceae; Deinococcus
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
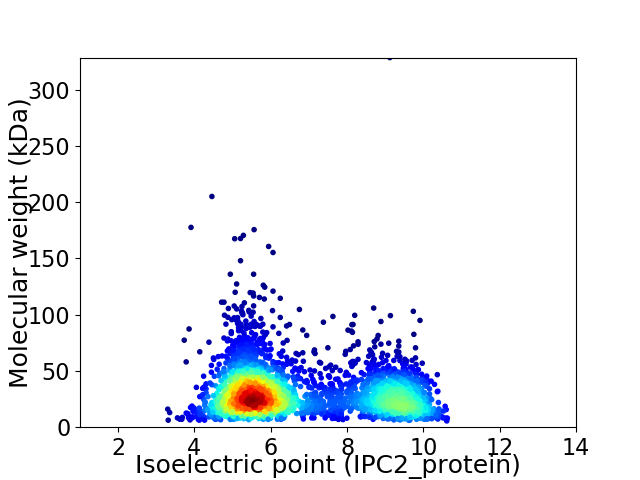
Virtual 2D-PAGE plot for 2935 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F7JS73|A0A0F7JS73_9DEIO DNA mismatch repair protein MutT OS=Deinococcus soli Cha et al. 2016 OX=1309411 GN=SY84_11125 PE=3 SV=1
MM1 pKa = 7.47KK2 pKa = 10.09KK3 pKa = 9.62SHH5 pKa = 7.11LIVLMAALAAGTAAAEE21 pKa = 4.35GTVAGTTIEE30 pKa = 4.21NTASASFQDD39 pKa = 4.0PANAGTTLTSNSNTVSTTVLPLPGFDD65 pKa = 3.57IVYY68 pKa = 10.46NDD70 pKa = 3.36STADD74 pKa = 3.38GNTISTTPILTTGAVPGQKK93 pKa = 8.54VTTGYY98 pKa = 10.81SVLNNGNVTLTVNLAADD115 pKa = 3.78TTGAASGQTVQYY127 pKa = 10.65YY128 pKa = 10.98LDD130 pKa = 4.05ANGDD134 pKa = 3.84GVADD138 pKa = 3.73STTPITSVIVPVDD151 pKa = 4.04DD152 pKa = 4.67PSTPADD158 pKa = 3.51EE159 pKa = 5.45GIVKK163 pKa = 10.01IVQVVTLPTDD173 pKa = 3.54PALITPSSIFGASPEE188 pKa = 4.13GSVTGTAGADD198 pKa = 3.45PLVTPGNGYY207 pKa = 8.48ATGTTVYY214 pKa = 10.7EE215 pKa = 4.42DD216 pKa = 3.35GKK218 pKa = 11.11AVNTDD223 pKa = 3.4LQFVRR228 pKa = 11.84ITVFAPALDD237 pKa = 4.19NNPNTNPSTPVDD249 pKa = 3.55SAGNPITDD257 pKa = 4.04PALVPPSQTVQIPTEE272 pKa = 4.38TTGKK276 pKa = 8.54TGDD279 pKa = 3.62ATPVVPTTGYY289 pKa = 7.7ITPGAPASDD298 pKa = 4.2PTPGGTPIVPNVAGDD313 pKa = 3.51IQIAYY318 pKa = 8.11PKK320 pKa = 10.56ADD322 pKa = 3.54TNTNPDD328 pKa = 3.14TVVFSNNLTNTSGATDD344 pKa = 3.49KK345 pKa = 11.33VQLFPALADD354 pKa = 3.74GTPDD358 pKa = 3.7PAYY361 pKa = 9.28TYY363 pKa = 10.97NAATGTFTNTTTGVVIRR380 pKa = 11.84FLDD383 pKa = 3.98PVTGAVILASTDD395 pKa = 3.43PNNPTTALYY404 pKa = 7.49PTVTVPTGSTVVYY417 pKa = 8.41RR418 pKa = 11.84TEE420 pKa = 3.69VTYY423 pKa = 10.46PDD425 pKa = 4.59PNDD428 pKa = 3.62SAVIAPVTVLIGADD442 pKa = 3.67SLKK445 pKa = 10.97DD446 pKa = 3.35AAIVSDD452 pKa = 3.76STTTNTILPPSAQFGDD468 pKa = 3.57ATGTLGAVSTPAPIQTINPNGTQSGVASPDD498 pKa = 3.25LTDD501 pKa = 3.4ATAVFPMDD509 pKa = 3.38VVNNGQYY516 pKa = 10.5NDD518 pKa = 4.03SYY520 pKa = 9.95TLSGSVTLNLVGGGTVTVPVLYY542 pKa = 9.93YY543 pKa = 10.95APDD546 pKa = 4.01GSLLPRR552 pKa = 11.84LSTDD556 pKa = 3.41PASPDD561 pKa = 3.13YY562 pKa = 10.85TKK564 pKa = 10.97FITPVIAPGTEE575 pKa = 3.78YY576 pKa = 10.79KK577 pKa = 10.35PYY579 pKa = 11.07AVVQVPAGTLTGDD592 pKa = 3.47YY593 pKa = 9.91TVTQNAVGNYY603 pKa = 6.78STIPMTDD610 pKa = 2.98ANNVIRR616 pKa = 11.84VAPDD620 pKa = 3.08GNVAVAKK627 pKa = 9.58FVAKK631 pKa = 10.44SGVTAASNPVNGINNPANFTATGATGAKK659 pKa = 9.42PGDD662 pKa = 4.54DD663 pKa = 2.58ITYY666 pKa = 10.51RR667 pKa = 11.84ITAKK671 pKa = 10.59NNYY674 pKa = 7.0NTTISNFFLSDD685 pKa = 3.42TVPANTLFKK694 pKa = 10.63SIQLLDD700 pKa = 3.44TSGTVISTNVIYY712 pKa = 10.64KK713 pKa = 9.87VGSGAWGTTAPAAGLANPSVIYY735 pKa = 10.3IALDD739 pKa = 3.53ANNDD743 pKa = 4.68LIPDD747 pKa = 4.44ALAPATTLTAEE758 pKa = 4.2FVVTVKK764 pKa = 10.91
MM1 pKa = 7.47KK2 pKa = 10.09KK3 pKa = 9.62SHH5 pKa = 7.11LIVLMAALAAGTAAAEE21 pKa = 4.35GTVAGTTIEE30 pKa = 4.21NTASASFQDD39 pKa = 4.0PANAGTTLTSNSNTVSTTVLPLPGFDD65 pKa = 3.57IVYY68 pKa = 10.46NDD70 pKa = 3.36STADD74 pKa = 3.38GNTISTTPILTTGAVPGQKK93 pKa = 8.54VTTGYY98 pKa = 10.81SVLNNGNVTLTVNLAADD115 pKa = 3.78TTGAASGQTVQYY127 pKa = 10.65YY128 pKa = 10.98LDD130 pKa = 4.05ANGDD134 pKa = 3.84GVADD138 pKa = 3.73STTPITSVIVPVDD151 pKa = 4.04DD152 pKa = 4.67PSTPADD158 pKa = 3.51EE159 pKa = 5.45GIVKK163 pKa = 10.01IVQVVTLPTDD173 pKa = 3.54PALITPSSIFGASPEE188 pKa = 4.13GSVTGTAGADD198 pKa = 3.45PLVTPGNGYY207 pKa = 8.48ATGTTVYY214 pKa = 10.7EE215 pKa = 4.42DD216 pKa = 3.35GKK218 pKa = 11.11AVNTDD223 pKa = 3.4LQFVRR228 pKa = 11.84ITVFAPALDD237 pKa = 4.19NNPNTNPSTPVDD249 pKa = 3.55SAGNPITDD257 pKa = 4.04PALVPPSQTVQIPTEE272 pKa = 4.38TTGKK276 pKa = 8.54TGDD279 pKa = 3.62ATPVVPTTGYY289 pKa = 7.7ITPGAPASDD298 pKa = 4.2PTPGGTPIVPNVAGDD313 pKa = 3.51IQIAYY318 pKa = 8.11PKK320 pKa = 10.56ADD322 pKa = 3.54TNTNPDD328 pKa = 3.14TVVFSNNLTNTSGATDD344 pKa = 3.49KK345 pKa = 11.33VQLFPALADD354 pKa = 3.74GTPDD358 pKa = 3.7PAYY361 pKa = 9.28TYY363 pKa = 10.97NAATGTFTNTTTGVVIRR380 pKa = 11.84FLDD383 pKa = 3.98PVTGAVILASTDD395 pKa = 3.43PNNPTTALYY404 pKa = 7.49PTVTVPTGSTVVYY417 pKa = 8.41RR418 pKa = 11.84TEE420 pKa = 3.69VTYY423 pKa = 10.46PDD425 pKa = 4.59PNDD428 pKa = 3.62SAVIAPVTVLIGADD442 pKa = 3.67SLKK445 pKa = 10.97DD446 pKa = 3.35AAIVSDD452 pKa = 3.76STTTNTILPPSAQFGDD468 pKa = 3.57ATGTLGAVSTPAPIQTINPNGTQSGVASPDD498 pKa = 3.25LTDD501 pKa = 3.4ATAVFPMDD509 pKa = 3.38VVNNGQYY516 pKa = 10.5NDD518 pKa = 4.03SYY520 pKa = 9.95TLSGSVTLNLVGGGTVTVPVLYY542 pKa = 9.93YY543 pKa = 10.95APDD546 pKa = 4.01GSLLPRR552 pKa = 11.84LSTDD556 pKa = 3.41PASPDD561 pKa = 3.13YY562 pKa = 10.85TKK564 pKa = 10.97FITPVIAPGTEE575 pKa = 3.78YY576 pKa = 10.79KK577 pKa = 10.35PYY579 pKa = 11.07AVVQVPAGTLTGDD592 pKa = 3.47YY593 pKa = 9.91TVTQNAVGNYY603 pKa = 6.78STIPMTDD610 pKa = 2.98ANNVIRR616 pKa = 11.84VAPDD620 pKa = 3.08GNVAVAKK627 pKa = 9.58FVAKK631 pKa = 10.44SGVTAASNPVNGINNPANFTATGATGAKK659 pKa = 9.42PGDD662 pKa = 4.54DD663 pKa = 2.58ITYY666 pKa = 10.51RR667 pKa = 11.84ITAKK671 pKa = 10.59NNYY674 pKa = 7.0NTTISNFFLSDD685 pKa = 3.42TVPANTLFKK694 pKa = 10.63SIQLLDD700 pKa = 3.44TSGTVISTNVIYY712 pKa = 10.64KK713 pKa = 9.87VGSGAWGTTAPAAGLANPSVIYY735 pKa = 10.3IALDD739 pKa = 3.53ANNDD743 pKa = 4.68LIPDD747 pKa = 4.44ALAPATTLTAEE758 pKa = 4.2FVVTVKK764 pKa = 10.91
Molecular weight: 77.34 kDa
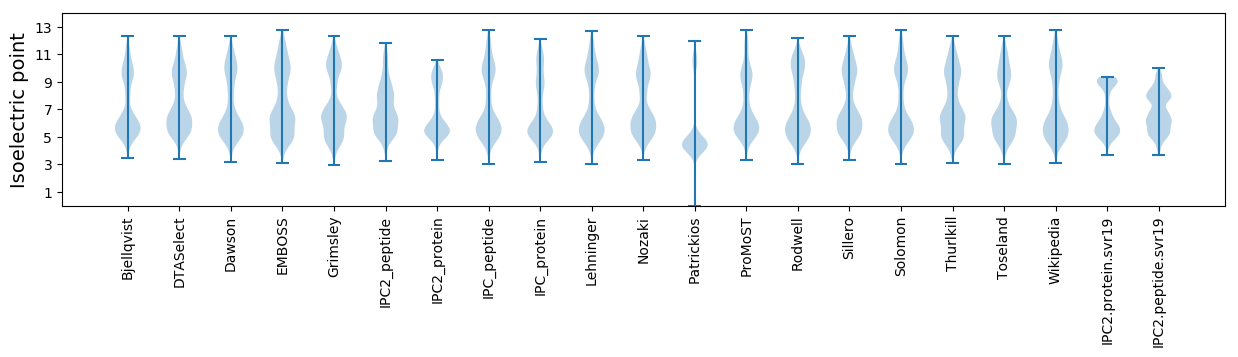
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F7JTQ2|A0A0F7JTQ2_9DEIO Peptide ABC transporter permease OS=Deinococcus soli Cha et al. 2016 OX=1309411 GN=SY84_14985 PE=3 SV=1
MM1 pKa = 7.14GASLAVLTAFLGEE14 pKa = 3.95VRR16 pKa = 11.84APAEE20 pKa = 4.22LLVGMIVTCGLAGAFRR36 pKa = 11.84PSWVALRR43 pKa = 11.84VGAGALAVLIAACLLTPVLRR63 pKa = 11.84IPLQSLTLRR72 pKa = 11.84EE73 pKa = 4.37PPVKK77 pKa = 10.42ADD79 pKa = 4.29AIVALGAGVHH89 pKa = 6.0CGSRR93 pKa = 11.84EE94 pKa = 3.99LEE96 pKa = 3.99ASAVARR102 pKa = 11.84LTGALALWRR111 pKa = 11.84AGYY114 pKa = 10.68APVVTVSEE122 pKa = 4.14QSGLIGPRR130 pKa = 11.84DD131 pKa = 4.0CPKK134 pKa = 10.13MSVLEE139 pKa = 4.29TEE141 pKa = 4.65LTRR144 pKa = 11.84ALYY147 pKa = 10.6GPGGPTLVTLKK158 pKa = 10.84NVTTTRR164 pKa = 11.84DD165 pKa = 2.82EE166 pKa = 4.11AARR169 pKa = 11.84VRR171 pKa = 11.84DD172 pKa = 3.7LTRR175 pKa = 11.84ARR177 pKa = 11.84GWTRR181 pKa = 11.84VLLVTSPSHH190 pKa = 5.82SRR192 pKa = 11.84RR193 pKa = 11.84AAALFRR199 pKa = 11.84SQGVTVVSVPAQEE212 pKa = 3.87WRR214 pKa = 11.84FDD216 pKa = 3.54QALTQPSDD224 pKa = 3.17RR225 pKa = 11.84LVALRR230 pKa = 11.84VLLYY234 pKa = 10.61EE235 pKa = 4.04GLSRR239 pKa = 11.84VKK241 pKa = 10.33AALGGTPEE249 pKa = 3.82RR250 pKa = 4.64
MM1 pKa = 7.14GASLAVLTAFLGEE14 pKa = 3.95VRR16 pKa = 11.84APAEE20 pKa = 4.22LLVGMIVTCGLAGAFRR36 pKa = 11.84PSWVALRR43 pKa = 11.84VGAGALAVLIAACLLTPVLRR63 pKa = 11.84IPLQSLTLRR72 pKa = 11.84EE73 pKa = 4.37PPVKK77 pKa = 10.42ADD79 pKa = 4.29AIVALGAGVHH89 pKa = 6.0CGSRR93 pKa = 11.84EE94 pKa = 3.99LEE96 pKa = 3.99ASAVARR102 pKa = 11.84LTGALALWRR111 pKa = 11.84AGYY114 pKa = 10.68APVVTVSEE122 pKa = 4.14QSGLIGPRR130 pKa = 11.84DD131 pKa = 4.0CPKK134 pKa = 10.13MSVLEE139 pKa = 4.29TEE141 pKa = 4.65LTRR144 pKa = 11.84ALYY147 pKa = 10.6GPGGPTLVTLKK158 pKa = 10.84NVTTTRR164 pKa = 11.84DD165 pKa = 2.82EE166 pKa = 4.11AARR169 pKa = 11.84VRR171 pKa = 11.84DD172 pKa = 3.7LTRR175 pKa = 11.84ARR177 pKa = 11.84GWTRR181 pKa = 11.84VLLVTSPSHH190 pKa = 5.82SRR192 pKa = 11.84RR193 pKa = 11.84AAALFRR199 pKa = 11.84SQGVTVVSVPAQEE212 pKa = 3.87WRR214 pKa = 11.84FDD216 pKa = 3.54QALTQPSDD224 pKa = 3.17RR225 pKa = 11.84LVALRR230 pKa = 11.84VLLYY234 pKa = 10.61EE235 pKa = 4.04GLSRR239 pKa = 11.84VKK241 pKa = 10.33AALGGTPEE249 pKa = 3.82RR250 pKa = 4.64
Molecular weight: 26.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
897889 |
47 |
3256 |
305.9 |
32.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.893 ± 0.061 | 0.53 ± 0.012 |
5.52 ± 0.033 | 5.027 ± 0.037 |
3.055 ± 0.029 | 9.347 ± 0.045 |
2.258 ± 0.033 | 3.389 ± 0.041 |
2.273 ± 0.038 | 11.617 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.827 ± 0.024 | 2.327 ± 0.034 |
5.952 ± 0.042 | 3.697 ± 0.033 |
7.524 ± 0.05 | 4.628 ± 0.033 |
6.538 ± 0.059 | 8.012 ± 0.043 |
1.376 ± 0.021 | 2.209 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
