
Nitrosomonas sp. Nm84
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Nitrosomonadaceae; Nitrosomonas; unclassified Nitrosomonas
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
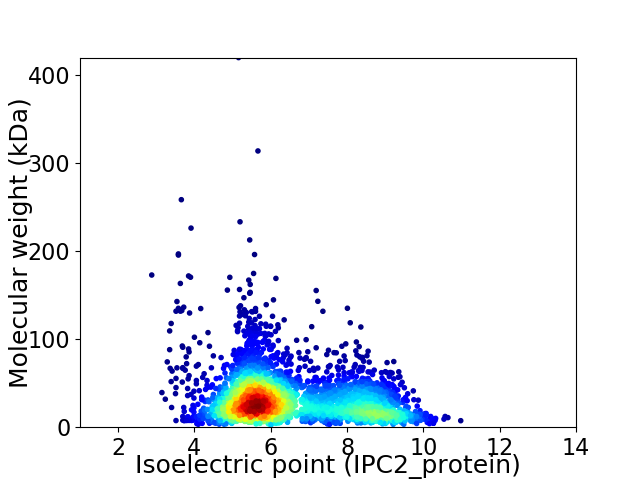
Virtual 2D-PAGE plot for 3167 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V3W3H2|A0A2V3W3H2_9PROT Uncharacterized protein OS=Nitrosomonas sp. Nm84 OX=200124 GN=C8R34_11047 PE=4 SV=1
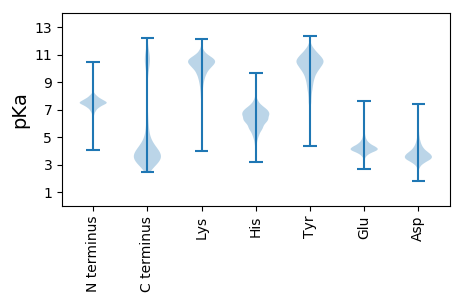
MM1 pKa = 7.74AIINGTSSSEE11 pKa = 4.36TVNGTTSDD19 pKa = 3.55DD20 pKa = 4.16TINGLGGSDD29 pKa = 3.67TLIGDD34 pKa = 4.75LGDD37 pKa = 3.61DD38 pKa = 3.67TYY40 pKa = 10.82EE41 pKa = 3.85YY42 pKa = 11.25SGIFSTTTLYY52 pKa = 10.29PYY54 pKa = 10.42TDD56 pKa = 3.13IVTEE60 pKa = 4.1LAGSGSDD67 pKa = 3.71TIHH70 pKa = 7.12ILTGSNPGDD79 pKa = 3.38IPASAVRR86 pKa = 11.84VQGGSASDD94 pKa = 4.65PNALWIIADD103 pKa = 4.55GYY105 pKa = 10.28GKK107 pKa = 10.16IYY109 pKa = 10.59LGNQLTAANVEE120 pKa = 4.27SLKK123 pKa = 10.6IGNNAPISLTSGINMTGSSSGEE145 pKa = 4.26TINGSAFNDD154 pKa = 4.37TINGMGGSDD163 pKa = 3.58TLIGGLGDD171 pKa = 3.49DD172 pKa = 3.9TYY174 pKa = 10.33EE175 pKa = 3.87YY176 pKa = 11.25SGIFSTTTLYY186 pKa = 10.29PYY188 pKa = 10.42TDD190 pKa = 3.13IVTEE194 pKa = 4.1LAGSGSDD201 pKa = 3.71TIHH204 pKa = 7.12ILTGSNPGDD213 pKa = 3.38IPASAVRR220 pKa = 11.84VQGGSTSDD228 pKa = 4.16PNSLWIIADD237 pKa = 4.4GYY239 pKa = 10.28GKK241 pKa = 10.16IYY243 pKa = 10.59LGNQLTAANVEE254 pKa = 4.27SLKK257 pKa = 10.6IGNNAPISLEE267 pKa = 3.79WLQIATGTSISLTSGLPMTGNINIQTISGSAYY299 pKa = 10.37NDD301 pKa = 3.49IINGKK306 pKa = 9.3GGNDD310 pKa = 3.43TLIGNLGNDD319 pKa = 3.52TYY321 pKa = 10.96IIGNAGVTITEE332 pKa = 4.38KK333 pKa = 10.92LNEE336 pKa = 4.23GTDD339 pKa = 3.79TVKK342 pKa = 11.01SSISYY347 pKa = 9.69ILPANVEE354 pKa = 4.14NLTLTGTLAINGTGNGSNNKK374 pKa = 8.47LTGNSAANQLKK385 pKa = 10.82GNGGNDD391 pKa = 3.5TLDD394 pKa = 3.39GKK396 pKa = 10.75AGNNTLTGGAGQDD409 pKa = 3.57TFKK412 pKa = 10.38LTSAGHH418 pKa = 6.49IDD420 pKa = 4.99AITDD424 pKa = 4.17FIAADD429 pKa = 3.56DD430 pKa = 4.5TIQLEE435 pKa = 4.4NAVFTALTTPGTLAAGQLKK454 pKa = 10.27IGTQALDD461 pKa = 3.49ANDD464 pKa = 3.21FVVYY468 pKa = 11.0NNVTGALLYY477 pKa = 10.93DD478 pKa = 4.48ADD480 pKa = 4.12ASGAAAAVQIATLSAGLALTNADD503 pKa = 3.47FVVII507 pKa = 4.58
MM1 pKa = 7.74AIINGTSSSEE11 pKa = 4.36TVNGTTSDD19 pKa = 3.55DD20 pKa = 4.16TINGLGGSDD29 pKa = 3.67TLIGDD34 pKa = 4.75LGDD37 pKa = 3.61DD38 pKa = 3.67TYY40 pKa = 10.82EE41 pKa = 3.85YY42 pKa = 11.25SGIFSTTTLYY52 pKa = 10.29PYY54 pKa = 10.42TDD56 pKa = 3.13IVTEE60 pKa = 4.1LAGSGSDD67 pKa = 3.71TIHH70 pKa = 7.12ILTGSNPGDD79 pKa = 3.38IPASAVRR86 pKa = 11.84VQGGSASDD94 pKa = 4.65PNALWIIADD103 pKa = 4.55GYY105 pKa = 10.28GKK107 pKa = 10.16IYY109 pKa = 10.59LGNQLTAANVEE120 pKa = 4.27SLKK123 pKa = 10.6IGNNAPISLTSGINMTGSSSGEE145 pKa = 4.26TINGSAFNDD154 pKa = 4.37TINGMGGSDD163 pKa = 3.58TLIGGLGDD171 pKa = 3.49DD172 pKa = 3.9TYY174 pKa = 10.33EE175 pKa = 3.87YY176 pKa = 11.25SGIFSTTTLYY186 pKa = 10.29PYY188 pKa = 10.42TDD190 pKa = 3.13IVTEE194 pKa = 4.1LAGSGSDD201 pKa = 3.71TIHH204 pKa = 7.12ILTGSNPGDD213 pKa = 3.38IPASAVRR220 pKa = 11.84VQGGSTSDD228 pKa = 4.16PNSLWIIADD237 pKa = 4.4GYY239 pKa = 10.28GKK241 pKa = 10.16IYY243 pKa = 10.59LGNQLTAANVEE254 pKa = 4.27SLKK257 pKa = 10.6IGNNAPISLEE267 pKa = 3.79WLQIATGTSISLTSGLPMTGNINIQTISGSAYY299 pKa = 10.37NDD301 pKa = 3.49IINGKK306 pKa = 9.3GGNDD310 pKa = 3.43TLIGNLGNDD319 pKa = 3.52TYY321 pKa = 10.96IIGNAGVTITEE332 pKa = 4.38KK333 pKa = 10.92LNEE336 pKa = 4.23GTDD339 pKa = 3.79TVKK342 pKa = 11.01SSISYY347 pKa = 9.69ILPANVEE354 pKa = 4.14NLTLTGTLAINGTGNGSNNKK374 pKa = 8.47LTGNSAANQLKK385 pKa = 10.82GNGGNDD391 pKa = 3.5TLDD394 pKa = 3.39GKK396 pKa = 10.75AGNNTLTGGAGQDD409 pKa = 3.57TFKK412 pKa = 10.38LTSAGHH418 pKa = 6.49IDD420 pKa = 4.99AITDD424 pKa = 4.17FIAADD429 pKa = 3.56DD430 pKa = 4.5TIQLEE435 pKa = 4.4NAVFTALTTPGTLAAGQLKK454 pKa = 10.27IGTQALDD461 pKa = 3.49ANDD464 pKa = 3.21FVVYY468 pKa = 11.0NNVTGALLYY477 pKa = 10.93DD478 pKa = 4.48ADD480 pKa = 4.12ASGAAAAVQIATLSAGLALTNADD503 pKa = 3.47FVVII507 pKa = 4.58
Molecular weight: 51.25 kDa
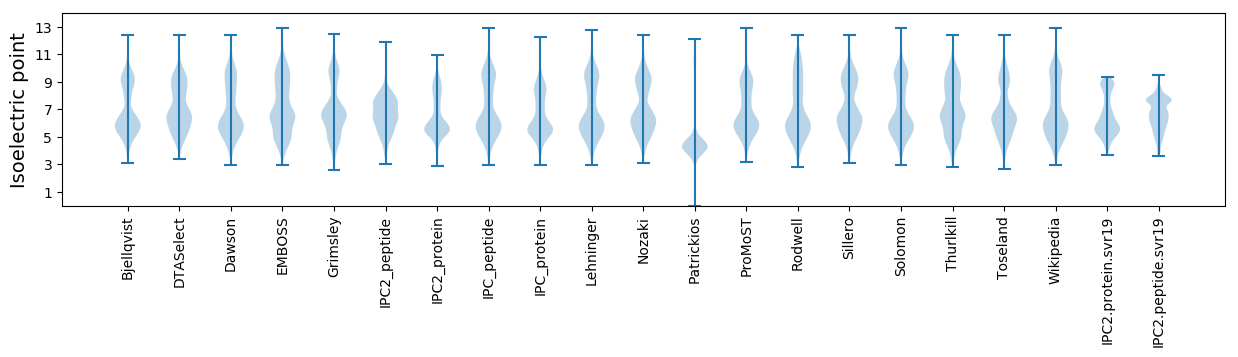
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V3WE06|A0A2V3WE06_9PROT Glutamate-1-semialdehyde 2 1-aminomutase OS=Nitrosomonas sp. Nm84 OX=200124 GN=hemL PE=3 SV=1
MM1 pKa = 8.08RR2 pKa = 11.84LRR4 pKa = 11.84SLCKK8 pKa = 10.08QRR10 pKa = 11.84VDD12 pKa = 3.25YY13 pKa = 11.1SDD15 pKa = 3.42AVRR18 pKa = 11.84FVKK21 pKa = 10.18KK22 pKa = 10.13CRR24 pKa = 11.84LIILAASIGQMMFDD38 pKa = 3.8VTRR41 pKa = 11.84EE42 pKa = 3.91MIALNRR48 pKa = 11.84FNLSFYY54 pKa = 10.87CRR56 pKa = 3.58
MM1 pKa = 8.08RR2 pKa = 11.84LRR4 pKa = 11.84SLCKK8 pKa = 10.08QRR10 pKa = 11.84VDD12 pKa = 3.25YY13 pKa = 11.1SDD15 pKa = 3.42AVRR18 pKa = 11.84FVKK21 pKa = 10.18KK22 pKa = 10.13CRR24 pKa = 11.84LIILAASIGQMMFDD38 pKa = 3.8VTRR41 pKa = 11.84EE42 pKa = 3.91MIALNRR48 pKa = 11.84FNLSFYY54 pKa = 10.87CRR56 pKa = 3.58
Molecular weight: 6.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
995109 |
27 |
3778 |
314.2 |
34.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.975 ± 0.052 | 0.952 ± 0.016 |
5.547 ± 0.054 | 5.686 ± 0.043 |
3.984 ± 0.028 | 7.09 ± 0.076 |
2.456 ± 0.028 | 6.951 ± 0.039 |
4.804 ± 0.053 | 10.328 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.45 ± 0.023 | 4.405 ± 0.046 |
4.155 ± 0.034 | 4.376 ± 0.038 |
5.214 ± 0.045 | 6.356 ± 0.047 |
5.602 ± 0.049 | 6.556 ± 0.04 |
1.231 ± 0.019 | 2.883 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
