
Giant house spider associated circular virus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus erati1
Average proteome isoelectric point is 7.43
Get precalculated fractions of proteins
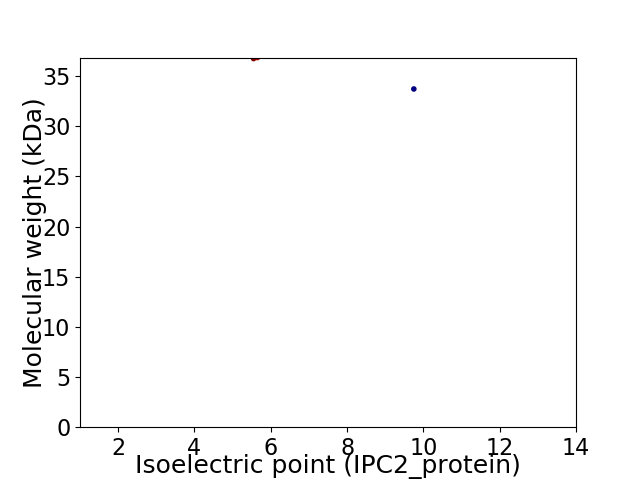
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BP83|A0A346BP83_9VIRU Putative capsid protein OS=Giant house spider associated circular virus 1 OX=2293288 PE=4 SV=1
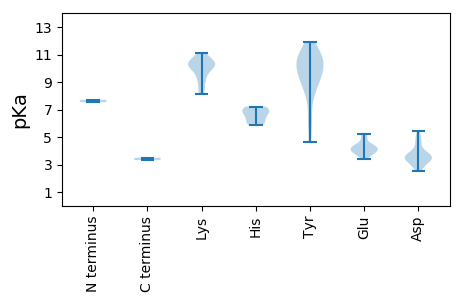
MM1 pKa = 7.71SFRR4 pKa = 11.84FNHH7 pKa = 6.52KK8 pKa = 10.25YY9 pKa = 10.79VLLTYY14 pKa = 8.91AQCGDD19 pKa = 3.65LDD21 pKa = 3.91GFAVMEE27 pKa = 4.95KK28 pKa = 9.92ISSLGAEE35 pKa = 4.25CIVARR40 pKa = 11.84EE41 pKa = 4.01HH42 pKa = 7.03HH43 pKa = 6.86EE44 pKa = 4.46DD45 pKa = 3.68GGVHH49 pKa = 6.12LHH51 pKa = 5.86VFCEE55 pKa = 4.49FEE57 pKa = 3.92RR58 pKa = 11.84KK59 pKa = 8.99FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84ADD66 pKa = 3.25VFDD69 pKa = 3.64VDD71 pKa = 4.58GVHH74 pKa = 7.22PNIEE78 pKa = 4.13PSKK81 pKa = 8.32GTPEE85 pKa = 3.79KK86 pKa = 10.85GYY88 pKa = 10.88DD89 pKa = 3.47YY90 pKa = 10.69AIKK93 pKa = 10.71DD94 pKa = 3.36GDD96 pKa = 4.31VICGGLARR104 pKa = 11.84PSARR108 pKa = 11.84PTTNGNTFDD117 pKa = 3.79KK118 pKa = 10.32WSQITSAEE126 pKa = 4.04SRR128 pKa = 11.84DD129 pKa = 3.9EE130 pKa = 4.2FWSLVHH136 pKa = 6.73EE137 pKa = 5.23LDD139 pKa = 4.49PKK141 pKa = 10.79SAACNHH147 pKa = 5.93SSLVKK152 pKa = 10.17YY153 pKa = 10.47CDD155 pKa = 2.5WKK157 pKa = 10.05YY158 pKa = 10.79RR159 pKa = 11.84EE160 pKa = 5.0RR161 pKa = 11.84IPDD164 pKa = 3.51YY165 pKa = 8.84KK166 pKa = 10.28TPAGISFDD174 pKa = 3.43SGGIDD179 pKa = 5.4GRR181 pKa = 11.84DD182 pKa = 2.96SWLQQSGIGLDD193 pKa = 3.53EE194 pKa = 4.49PLVGKK199 pKa = 10.56LSYY202 pKa = 9.74TSQGGIPPTGKK213 pKa = 8.15TLWARR218 pKa = 11.84SLGAHH223 pKa = 7.15IYY225 pKa = 9.06CVGMISGDD233 pKa = 3.51EE234 pKa = 4.27CIKK237 pKa = 10.31TDD239 pKa = 2.81VDD241 pKa = 3.7YY242 pKa = 11.49AIFDD246 pKa = 5.02DD247 pKa = 3.61IRR249 pKa = 11.84GGIKK253 pKa = 9.99FFPSFKK259 pKa = 10.22EE260 pKa = 3.81WLGCQQWITVKK271 pKa = 10.48RR272 pKa = 11.84LYY274 pKa = 10.44RR275 pKa = 11.84EE276 pKa = 3.79PKK278 pKa = 9.24LRR280 pKa = 11.84LWGKK284 pKa = 8.42PSIWISNTDD293 pKa = 3.32PRR295 pKa = 11.84NEE297 pKa = 3.86MLQSDD302 pKa = 4.98VEE304 pKa = 4.35WMNLNCTFIEE314 pKa = 4.22VNSAIFHH321 pKa = 6.9ANIQQ325 pKa = 3.5
MM1 pKa = 7.71SFRR4 pKa = 11.84FNHH7 pKa = 6.52KK8 pKa = 10.25YY9 pKa = 10.79VLLTYY14 pKa = 8.91AQCGDD19 pKa = 3.65LDD21 pKa = 3.91GFAVMEE27 pKa = 4.95KK28 pKa = 9.92ISSLGAEE35 pKa = 4.25CIVARR40 pKa = 11.84EE41 pKa = 4.01HH42 pKa = 7.03HH43 pKa = 6.86EE44 pKa = 4.46DD45 pKa = 3.68GGVHH49 pKa = 6.12LHH51 pKa = 5.86VFCEE55 pKa = 4.49FEE57 pKa = 3.92RR58 pKa = 11.84KK59 pKa = 8.99FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84ADD66 pKa = 3.25VFDD69 pKa = 3.64VDD71 pKa = 4.58GVHH74 pKa = 7.22PNIEE78 pKa = 4.13PSKK81 pKa = 8.32GTPEE85 pKa = 3.79KK86 pKa = 10.85GYY88 pKa = 10.88DD89 pKa = 3.47YY90 pKa = 10.69AIKK93 pKa = 10.71DD94 pKa = 3.36GDD96 pKa = 4.31VICGGLARR104 pKa = 11.84PSARR108 pKa = 11.84PTTNGNTFDD117 pKa = 3.79KK118 pKa = 10.32WSQITSAEE126 pKa = 4.04SRR128 pKa = 11.84DD129 pKa = 3.9EE130 pKa = 4.2FWSLVHH136 pKa = 6.73EE137 pKa = 5.23LDD139 pKa = 4.49PKK141 pKa = 10.79SAACNHH147 pKa = 5.93SSLVKK152 pKa = 10.17YY153 pKa = 10.47CDD155 pKa = 2.5WKK157 pKa = 10.05YY158 pKa = 10.79RR159 pKa = 11.84EE160 pKa = 5.0RR161 pKa = 11.84IPDD164 pKa = 3.51YY165 pKa = 8.84KK166 pKa = 10.28TPAGISFDD174 pKa = 3.43SGGIDD179 pKa = 5.4GRR181 pKa = 11.84DD182 pKa = 2.96SWLQQSGIGLDD193 pKa = 3.53EE194 pKa = 4.49PLVGKK199 pKa = 10.56LSYY202 pKa = 9.74TSQGGIPPTGKK213 pKa = 8.15TLWARR218 pKa = 11.84SLGAHH223 pKa = 7.15IYY225 pKa = 9.06CVGMISGDD233 pKa = 3.51EE234 pKa = 4.27CIKK237 pKa = 10.31TDD239 pKa = 2.81VDD241 pKa = 3.7YY242 pKa = 11.49AIFDD246 pKa = 5.02DD247 pKa = 3.61IRR249 pKa = 11.84GGIKK253 pKa = 9.99FFPSFKK259 pKa = 10.22EE260 pKa = 3.81WLGCQQWITVKK271 pKa = 10.48RR272 pKa = 11.84LYY274 pKa = 10.44RR275 pKa = 11.84EE276 pKa = 3.79PKK278 pKa = 9.24LRR280 pKa = 11.84LWGKK284 pKa = 8.42PSIWISNTDD293 pKa = 3.32PRR295 pKa = 11.84NEE297 pKa = 3.86MLQSDD302 pKa = 4.98VEE304 pKa = 4.35WMNLNCTFIEE314 pKa = 4.22VNSAIFHH321 pKa = 6.9ANIQQ325 pKa = 3.5
Molecular weight: 36.74 kDa
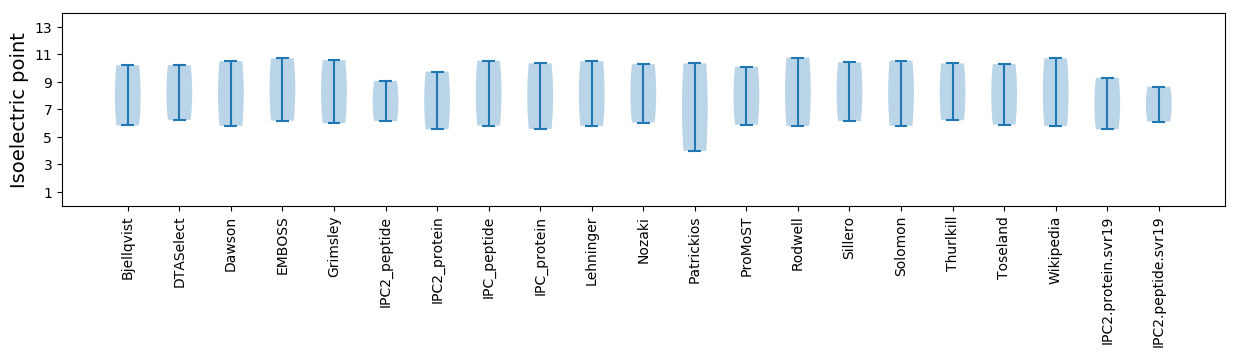
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BP83|A0A346BP83_9VIRU Putative capsid protein OS=Giant house spider associated circular virus 1 OX=2293288 PE=4 SV=1
MM1 pKa = 7.53GKK3 pKa = 8.41YY4 pKa = 6.03TTSKK8 pKa = 10.28SSGRR12 pKa = 11.84YY13 pKa = 4.63GTKK16 pKa = 9.59RR17 pKa = 11.84YY18 pKa = 9.89ARR20 pKa = 11.84RR21 pKa = 11.84SRR23 pKa = 11.84TTIKK27 pKa = 9.93RR28 pKa = 11.84RR29 pKa = 11.84YY30 pKa = 6.53PARR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 9.17RR36 pKa = 11.84AYY38 pKa = 8.37TKK40 pKa = 10.21RR41 pKa = 11.84PRR43 pKa = 11.84PMSRR47 pKa = 11.84KK48 pKa = 9.59RR49 pKa = 11.84ILNVSTTKK57 pKa = 10.52KK58 pKa = 10.2RR59 pKa = 11.84DD60 pKa = 3.25TLMSYY65 pKa = 10.89DD66 pKa = 3.44VARR69 pKa = 11.84SQNGPAIVSSGSVEE83 pKa = 3.51RR84 pKa = 11.84WTTNLLNYY92 pKa = 10.17LPTSFSSADD101 pKa = 3.22DD102 pKa = 3.78DD103 pKa = 4.61YY104 pKa = 11.89RR105 pKa = 11.84RR106 pKa = 11.84TKK108 pKa = 10.32NNVYY112 pKa = 10.14FRR114 pKa = 11.84GAKK117 pKa = 9.58LAWNFDD123 pKa = 3.96MSDD126 pKa = 3.24SSPWEE131 pKa = 3.41IRR133 pKa = 11.84MIAFWGSDD141 pKa = 3.31GTITANPQSRR151 pKa = 11.84FYY153 pKa = 9.1TTITPTGGSPYY164 pKa = 10.0QVNGLLPVLPGSVPDD179 pKa = 3.87LRR181 pKa = 11.84MTLFQGTEE189 pKa = 3.85GTDD192 pKa = 2.67WFDD195 pKa = 5.04AMIAPLDD202 pKa = 3.51VRR204 pKa = 11.84RR205 pKa = 11.84NKK207 pKa = 10.25IIIDD211 pKa = 3.17KK212 pKa = 10.87RR213 pKa = 11.84MTLRR217 pKa = 11.84SGNDD221 pKa = 2.9VGSFKK226 pKa = 10.86RR227 pKa = 11.84MNLWVPANKK236 pKa = 9.09MVRR239 pKa = 11.84FNNDD243 pKa = 2.6EE244 pKa = 4.25NGSDD248 pKa = 3.52VDD250 pKa = 3.89KK251 pKa = 11.11TSGIPSGLYY260 pKa = 10.62SPDD263 pKa = 3.01CLYY266 pKa = 11.23YY267 pKa = 9.44VTLYY271 pKa = 9.62KK272 pKa = 10.37QRR274 pKa = 11.84GAADD278 pKa = 3.32TAAQLLVSCNATVYY292 pKa = 8.05WHH294 pKa = 6.33EE295 pKa = 4.12RR296 pKa = 3.32
MM1 pKa = 7.53GKK3 pKa = 8.41YY4 pKa = 6.03TTSKK8 pKa = 10.28SSGRR12 pKa = 11.84YY13 pKa = 4.63GTKK16 pKa = 9.59RR17 pKa = 11.84YY18 pKa = 9.89ARR20 pKa = 11.84RR21 pKa = 11.84SRR23 pKa = 11.84TTIKK27 pKa = 9.93RR28 pKa = 11.84RR29 pKa = 11.84YY30 pKa = 6.53PARR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 9.17RR36 pKa = 11.84AYY38 pKa = 8.37TKK40 pKa = 10.21RR41 pKa = 11.84PRR43 pKa = 11.84PMSRR47 pKa = 11.84KK48 pKa = 9.59RR49 pKa = 11.84ILNVSTTKK57 pKa = 10.52KK58 pKa = 10.2RR59 pKa = 11.84DD60 pKa = 3.25TLMSYY65 pKa = 10.89DD66 pKa = 3.44VARR69 pKa = 11.84SQNGPAIVSSGSVEE83 pKa = 3.51RR84 pKa = 11.84WTTNLLNYY92 pKa = 10.17LPTSFSSADD101 pKa = 3.22DD102 pKa = 3.78DD103 pKa = 4.61YY104 pKa = 11.89RR105 pKa = 11.84RR106 pKa = 11.84TKK108 pKa = 10.32NNVYY112 pKa = 10.14FRR114 pKa = 11.84GAKK117 pKa = 9.58LAWNFDD123 pKa = 3.96MSDD126 pKa = 3.24SSPWEE131 pKa = 3.41IRR133 pKa = 11.84MIAFWGSDD141 pKa = 3.31GTITANPQSRR151 pKa = 11.84FYY153 pKa = 9.1TTITPTGGSPYY164 pKa = 10.0QVNGLLPVLPGSVPDD179 pKa = 3.87LRR181 pKa = 11.84MTLFQGTEE189 pKa = 3.85GTDD192 pKa = 2.67WFDD195 pKa = 5.04AMIAPLDD202 pKa = 3.51VRR204 pKa = 11.84RR205 pKa = 11.84NKK207 pKa = 10.25IIIDD211 pKa = 3.17KK212 pKa = 10.87RR213 pKa = 11.84MTLRR217 pKa = 11.84SGNDD221 pKa = 2.9VGSFKK226 pKa = 10.86RR227 pKa = 11.84MNLWVPANKK236 pKa = 9.09MVRR239 pKa = 11.84FNNDD243 pKa = 2.6EE244 pKa = 4.25NGSDD248 pKa = 3.52VDD250 pKa = 3.89KK251 pKa = 11.11TSGIPSGLYY260 pKa = 10.62SPDD263 pKa = 3.01CLYY266 pKa = 11.23YY267 pKa = 9.44VTLYY271 pKa = 9.62KK272 pKa = 10.37QRR274 pKa = 11.84GAADD278 pKa = 3.32TAAQLLVSCNATVYY292 pKa = 8.05WHH294 pKa = 6.33EE295 pKa = 4.12RR296 pKa = 3.32
Molecular weight: 33.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
621 |
296 |
325 |
310.5 |
35.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.636 ± 0.269 | 1.932 ± 0.759 |
7.085 ± 0.402 | 3.865 ± 1.314 |
4.026 ± 0.595 | 7.89 ± 0.685 |
1.771 ± 0.866 | 5.636 ± 0.955 |
5.797 ± 0.033 | 6.441 ± 0.013 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.415 ± 0.582 | 4.509 ± 0.746 |
4.992 ± 0.25 | 2.415 ± 0.235 |
7.89 ± 1.56 | 8.696 ± 0.461 |
6.602 ± 1.522 | 5.314 ± 0.055 |
2.738 ± 0.225 | 4.348 ± 0.639 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
