
Apple chlorotic leaf spot virus (isolate plum P863) (ACLSV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Trivirinae; Trichovirus; Apple chlorotic leaf spot virus
Average proteome isoelectric point is 7.25
Get precalculated fractions of proteins
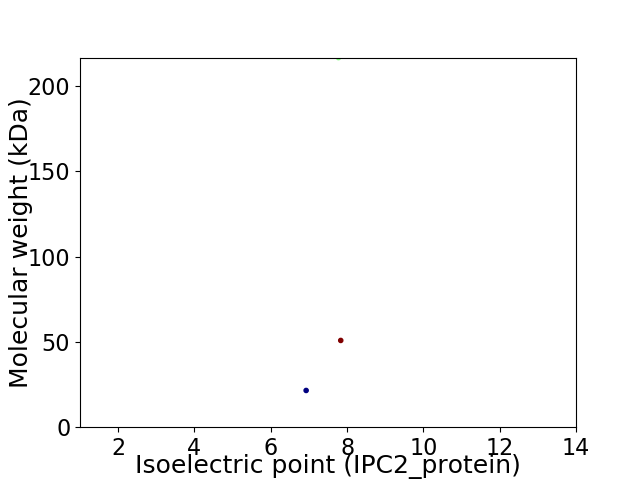
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P27738|RDRP_ACLSP RNA-directed RNA polymerase OS=Apple chlorotic leaf spot virus (isolate plum P863) OX=73473 PE=4 SV=1
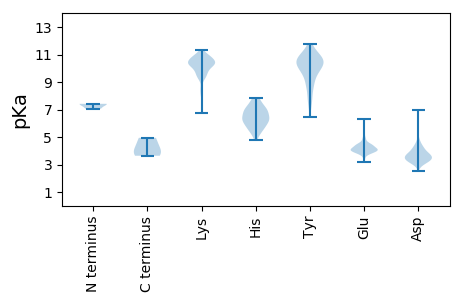
MM1 pKa = 7.42AAVLNLQLKK10 pKa = 9.92VDD12 pKa = 4.05ASLKK16 pKa = 10.59AFLGAEE22 pKa = 3.85NRR24 pKa = 11.84PLHH27 pKa = 6.12GKK29 pKa = 7.87TGATLEE35 pKa = 4.37QILEE39 pKa = 4.41SIFANIAIQGTSEE52 pKa = 3.77QTEE55 pKa = 4.05FLDD58 pKa = 4.33LVVEE62 pKa = 4.51VKK64 pKa = 10.87SMEE67 pKa = 4.3DD68 pKa = 3.25QSVLGSYY75 pKa = 8.65NLKK78 pKa = 10.49EE79 pKa = 4.07VVNLIKK85 pKa = 10.66AFKK88 pKa = 8.02TTSSDD93 pKa = 3.42PNINKK98 pKa = 6.75MTFRR102 pKa = 11.84QVCEE106 pKa = 3.82AFAPEE111 pKa = 4.08ARR113 pKa = 11.84NGLVKK118 pKa = 10.76LKK120 pKa = 10.82YY121 pKa = 10.13KK122 pKa = 10.81GVFTNLFTTMPEE134 pKa = 4.02VGSKK138 pKa = 10.57YY139 pKa = 9.76PEE141 pKa = 4.09LMFDD145 pKa = 4.06FNKK148 pKa = 9.68GLNMFIMNKK157 pKa = 9.41AQQKK161 pKa = 10.48VITNMNRR168 pKa = 11.84RR169 pKa = 11.84LLQTEE174 pKa = 4.41FAKK177 pKa = 10.82SEE179 pKa = 4.12NEE181 pKa = 3.8AKK183 pKa = 10.48LSSVSTDD190 pKa = 2.99LCII193 pKa = 4.94
MM1 pKa = 7.42AAVLNLQLKK10 pKa = 9.92VDD12 pKa = 4.05ASLKK16 pKa = 10.59AFLGAEE22 pKa = 3.85NRR24 pKa = 11.84PLHH27 pKa = 6.12GKK29 pKa = 7.87TGATLEE35 pKa = 4.37QILEE39 pKa = 4.41SIFANIAIQGTSEE52 pKa = 3.77QTEE55 pKa = 4.05FLDD58 pKa = 4.33LVVEE62 pKa = 4.51VKK64 pKa = 10.87SMEE67 pKa = 4.3DD68 pKa = 3.25QSVLGSYY75 pKa = 8.65NLKK78 pKa = 10.49EE79 pKa = 4.07VVNLIKK85 pKa = 10.66AFKK88 pKa = 8.02TTSSDD93 pKa = 3.42PNINKK98 pKa = 6.75MTFRR102 pKa = 11.84QVCEE106 pKa = 3.82AFAPEE111 pKa = 4.08ARR113 pKa = 11.84NGLVKK118 pKa = 10.76LKK120 pKa = 10.82YY121 pKa = 10.13KK122 pKa = 10.81GVFTNLFTTMPEE134 pKa = 4.02VGSKK138 pKa = 10.57YY139 pKa = 9.76PEE141 pKa = 4.09LMFDD145 pKa = 4.06FNKK148 pKa = 9.68GLNMFIMNKK157 pKa = 9.41AQQKK161 pKa = 10.48VITNMNRR168 pKa = 11.84RR169 pKa = 11.84LLQTEE174 pKa = 4.41FAKK177 pKa = 10.82SEE179 pKa = 4.12NEE181 pKa = 3.8AKK183 pKa = 10.48LSSVSTDD190 pKa = 2.99LCII193 pKa = 4.94
Molecular weight: 21.53 kDa
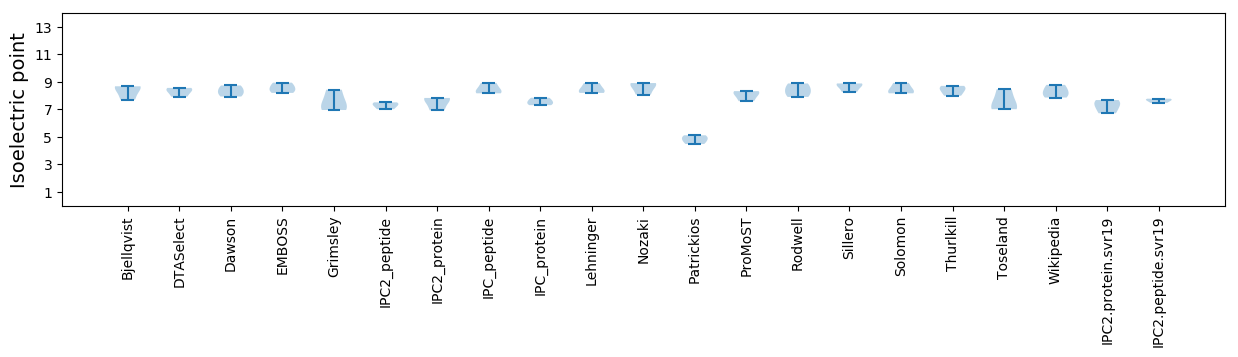
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P27739|MP_ACLSP Putative movement protein OS=Apple chlorotic leaf spot virus (isolate plum P863) OX=73473 PE=3 SV=1
MM1 pKa = 7.06ATMIRR6 pKa = 11.84GHH8 pKa = 6.43RR9 pKa = 11.84LRR11 pKa = 11.84IAEE14 pKa = 4.07GDD16 pKa = 3.53IPIAGVKK23 pKa = 9.81SSRR26 pKa = 11.84IYY28 pKa = 11.17SDD30 pKa = 3.19ISPFKK35 pKa = 10.32KK36 pKa = 10.55ASDD39 pKa = 4.81LMIHH43 pKa = 5.83WNEE46 pKa = 3.84FVFKK50 pKa = 10.88VMPEE54 pKa = 4.42DD55 pKa = 3.29IAGDD59 pKa = 4.15GFRR62 pKa = 11.84LASIPVIPSSEE73 pKa = 4.05VQAVLRR79 pKa = 11.84KK80 pKa = 9.69RR81 pKa = 11.84EE82 pKa = 4.1STNYY86 pKa = 7.76VHH88 pKa = 7.3WGALSISIDD97 pKa = 3.16ALFRR101 pKa = 11.84KK102 pKa = 9.49NAGVSGWCYY111 pKa = 10.45VYY113 pKa = 11.0DD114 pKa = 3.98NRR116 pKa = 11.84WEE118 pKa = 4.18TFEE121 pKa = 4.38QAMLQKK127 pKa = 10.27FHH129 pKa = 7.29FNLDD133 pKa = 3.51SGSATLVTSPNFPVSLDD150 pKa = 3.76DD151 pKa = 4.79PGLSNSISVAVMFEE165 pKa = 3.7NLNFKK170 pKa = 10.62FEE172 pKa = 4.6SYY174 pKa = 9.85PISVRR179 pKa = 11.84VGNMCRR185 pKa = 11.84FFDD188 pKa = 4.15SFLSSVKK195 pKa = 10.83NKK197 pKa = 9.81VDD199 pKa = 4.18SNFLLEE205 pKa = 4.93ASNADD210 pKa = 3.83PLGVGAFGFEE220 pKa = 3.93QDD222 pKa = 4.16DD223 pKa = 3.85QVSEE227 pKa = 4.25LFNYY231 pKa = 8.89IQTVPTQAIKK241 pKa = 10.43FRR243 pKa = 11.84EE244 pKa = 4.01HH245 pKa = 7.31EE246 pKa = 4.1IPKK249 pKa = 10.41GFLGMMGKK257 pKa = 9.94KK258 pKa = 9.57KK259 pKa = 10.19IKK261 pKa = 10.29SFEE264 pKa = 4.01FASGSKK270 pKa = 10.26GMEE273 pKa = 3.17RR274 pKa = 11.84RR275 pKa = 11.84KK276 pKa = 10.01PNRR279 pKa = 11.84GKK281 pKa = 10.62QIDD284 pKa = 3.53RR285 pKa = 11.84SFSQRR290 pKa = 11.84AVPGFRR296 pKa = 11.84SQNEE300 pKa = 4.23KK301 pKa = 10.61VEE303 pKa = 4.16HH304 pKa = 6.17QGLSTDD310 pKa = 3.31SDD312 pKa = 3.95FEE314 pKa = 4.14NFLRR318 pKa = 11.84NKK320 pKa = 9.58RR321 pKa = 11.84GNKK324 pKa = 9.97AGVKK328 pKa = 8.55STASEE333 pKa = 4.33GSSVDD338 pKa = 3.67NISSRR343 pKa = 11.84EE344 pKa = 3.83FQFARR349 pKa = 11.84QNQAKK354 pKa = 9.3EE355 pKa = 4.42DD356 pKa = 3.86GSSSEE361 pKa = 4.17FAAQGGRR368 pKa = 11.84KK369 pKa = 9.19SKK371 pKa = 10.75GISGRR376 pKa = 11.84RR377 pKa = 11.84KK378 pKa = 8.27QTSSWKK384 pKa = 10.28DD385 pKa = 3.04RR386 pKa = 11.84GNPGTDD392 pKa = 2.55TGVHH396 pKa = 5.57LRR398 pKa = 11.84EE399 pKa = 4.5HH400 pKa = 6.98SDD402 pKa = 3.4PGNVRR407 pKa = 11.84ADD409 pKa = 3.64GVSGPSGGSEE419 pKa = 3.8INGGSISPRR428 pKa = 11.84VLQPEE433 pKa = 4.53GSGQLDD439 pKa = 3.41QSFQDD444 pKa = 3.65YY445 pKa = 10.98LFGPEE450 pKa = 4.28HH451 pKa = 5.68QQNDD455 pKa = 3.66IPSGLL460 pKa = 3.64
MM1 pKa = 7.06ATMIRR6 pKa = 11.84GHH8 pKa = 6.43RR9 pKa = 11.84LRR11 pKa = 11.84IAEE14 pKa = 4.07GDD16 pKa = 3.53IPIAGVKK23 pKa = 9.81SSRR26 pKa = 11.84IYY28 pKa = 11.17SDD30 pKa = 3.19ISPFKK35 pKa = 10.32KK36 pKa = 10.55ASDD39 pKa = 4.81LMIHH43 pKa = 5.83WNEE46 pKa = 3.84FVFKK50 pKa = 10.88VMPEE54 pKa = 4.42DD55 pKa = 3.29IAGDD59 pKa = 4.15GFRR62 pKa = 11.84LASIPVIPSSEE73 pKa = 4.05VQAVLRR79 pKa = 11.84KK80 pKa = 9.69RR81 pKa = 11.84EE82 pKa = 4.1STNYY86 pKa = 7.76VHH88 pKa = 7.3WGALSISIDD97 pKa = 3.16ALFRR101 pKa = 11.84KK102 pKa = 9.49NAGVSGWCYY111 pKa = 10.45VYY113 pKa = 11.0DD114 pKa = 3.98NRR116 pKa = 11.84WEE118 pKa = 4.18TFEE121 pKa = 4.38QAMLQKK127 pKa = 10.27FHH129 pKa = 7.29FNLDD133 pKa = 3.51SGSATLVTSPNFPVSLDD150 pKa = 3.76DD151 pKa = 4.79PGLSNSISVAVMFEE165 pKa = 3.7NLNFKK170 pKa = 10.62FEE172 pKa = 4.6SYY174 pKa = 9.85PISVRR179 pKa = 11.84VGNMCRR185 pKa = 11.84FFDD188 pKa = 4.15SFLSSVKK195 pKa = 10.83NKK197 pKa = 9.81VDD199 pKa = 4.18SNFLLEE205 pKa = 4.93ASNADD210 pKa = 3.83PLGVGAFGFEE220 pKa = 3.93QDD222 pKa = 4.16DD223 pKa = 3.85QVSEE227 pKa = 4.25LFNYY231 pKa = 8.89IQTVPTQAIKK241 pKa = 10.43FRR243 pKa = 11.84EE244 pKa = 4.01HH245 pKa = 7.31EE246 pKa = 4.1IPKK249 pKa = 10.41GFLGMMGKK257 pKa = 9.94KK258 pKa = 9.57KK259 pKa = 10.19IKK261 pKa = 10.29SFEE264 pKa = 4.01FASGSKK270 pKa = 10.26GMEE273 pKa = 3.17RR274 pKa = 11.84RR275 pKa = 11.84KK276 pKa = 10.01PNRR279 pKa = 11.84GKK281 pKa = 10.62QIDD284 pKa = 3.53RR285 pKa = 11.84SFSQRR290 pKa = 11.84AVPGFRR296 pKa = 11.84SQNEE300 pKa = 4.23KK301 pKa = 10.61VEE303 pKa = 4.16HH304 pKa = 6.17QGLSTDD310 pKa = 3.31SDD312 pKa = 3.95FEE314 pKa = 4.14NFLRR318 pKa = 11.84NKK320 pKa = 9.58RR321 pKa = 11.84GNKK324 pKa = 9.97AGVKK328 pKa = 8.55STASEE333 pKa = 4.33GSSVDD338 pKa = 3.67NISSRR343 pKa = 11.84EE344 pKa = 3.83FQFARR349 pKa = 11.84QNQAKK354 pKa = 9.3EE355 pKa = 4.42DD356 pKa = 3.86GSSSEE361 pKa = 4.17FAAQGGRR368 pKa = 11.84KK369 pKa = 9.19SKK371 pKa = 10.75GISGRR376 pKa = 11.84RR377 pKa = 11.84KK378 pKa = 8.27QTSSWKK384 pKa = 10.28DD385 pKa = 3.04RR386 pKa = 11.84GNPGTDD392 pKa = 2.55TGVHH396 pKa = 5.57LRR398 pKa = 11.84EE399 pKa = 4.5HH400 pKa = 6.98SDD402 pKa = 3.4PGNVRR407 pKa = 11.84ADD409 pKa = 3.64GVSGPSGGSEE419 pKa = 3.8INGGSISPRR428 pKa = 11.84VLQPEE433 pKa = 4.53GSGQLDD439 pKa = 3.41QSFQDD444 pKa = 3.65YY445 pKa = 10.98LFGPEE450 pKa = 4.28HH451 pKa = 5.68QQNDD455 pKa = 3.66IPSGLL460 pKa = 3.64
Molecular weight: 50.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2537 |
193 |
1884 |
845.7 |
96.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.124 ± 0.786 | 2.05 ± 0.757 |
5.558 ± 0.646 | 7.174 ± 0.518 |
5.952 ± 0.426 | 6.543 ± 1.22 |
2.444 ± 0.592 | 6.07 ± 0.563 |
8.08 ± 0.788 | 8.79 ± 1.39 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.838 ± 0.396 | 4.493 ± 1.032 |
3.666 ± 0.415 | 3.193 ± 0.821 |
5.873 ± 0.866 | 8.869 ± 1.419 |
3.508 ± 0.871 | 5.794 ± 0.434 |
1.064 ± 0.285 | 2.917 ± 0.733 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
