
Yellowstone lake phycodnavirus 3
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Algavirales; Phycodnaviridae; Prasinovirus; unclassified Prasinovirus
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
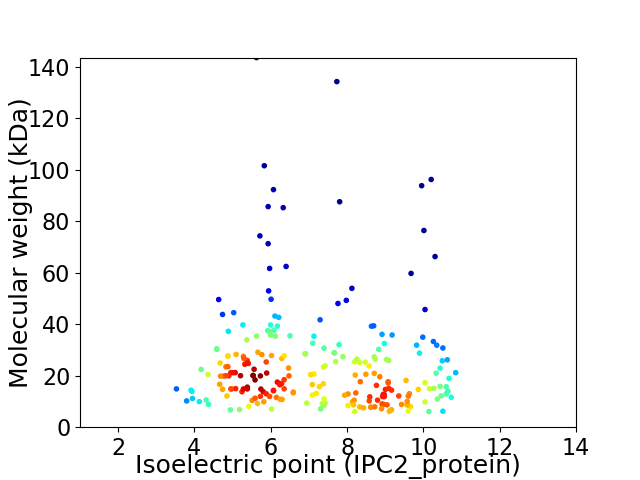
Virtual 2D-PAGE plot for 236 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P0YNP1|A0A0P0YNP1_9PHYC RING-type domain-containing protein OS=Yellowstone lake phycodnavirus 3 OX=1586715 PE=4 SV=1
MM1 pKa = 7.43SPNAYY6 pKa = 9.05VHH8 pKa = 6.67WDD10 pKa = 3.36DD11 pKa = 4.75EE12 pKa = 4.31KK13 pKa = 11.31RR14 pKa = 11.84LLFGFQGEE22 pKa = 4.58AEE24 pKa = 4.17HH25 pKa = 6.41EE26 pKa = 4.34KK27 pKa = 10.72PVFTWTYY34 pKa = 8.67STFEE38 pKa = 4.09EE39 pKa = 4.9YY40 pKa = 10.95LNEE43 pKa = 4.17ACTPSMYY50 pKa = 10.81QFMANYY56 pKa = 9.27IIEE59 pKa = 4.93EE60 pKa = 3.92ISKK63 pKa = 11.06DD64 pKa = 3.57EE65 pKa = 4.34LYY67 pKa = 10.68EE68 pKa = 3.95LQGGDD73 pKa = 3.75YY74 pKa = 10.93CPGRR78 pKa = 11.84LEE80 pKa = 4.15EE81 pKa = 4.36EE82 pKa = 4.13AVDD85 pKa = 4.92AYY87 pKa = 11.16FDD89 pKa = 3.96LPINEE94 pKa = 4.75RR95 pKa = 11.84IEE97 pKa = 4.03LHH99 pKa = 5.51NQEE102 pKa = 5.13LLNLYY107 pKa = 10.29AEE109 pKa = 4.79RR110 pKa = 11.84DD111 pKa = 3.41RR112 pKa = 11.84ATGKK116 pKa = 9.72EE117 pKa = 3.68AAFIDD122 pKa = 3.93ACLDD126 pKa = 3.39DD127 pKa = 4.99HH128 pKa = 7.53CPFDD132 pKa = 4.3VDD134 pKa = 3.74SPIYY138 pKa = 10.65VEE140 pKa = 4.09YY141 pKa = 10.63CQWCRR146 pKa = 11.84KK147 pKa = 7.24QTEE150 pKa = 3.84KK151 pKa = 7.96WTKK154 pKa = 9.63IVARR158 pKa = 11.84LADD161 pKa = 4.87EE162 pKa = 3.95IHH164 pKa = 6.87EE165 pKa = 4.37EE166 pKa = 4.25EE167 pKa = 4.79DD168 pKa = 3.19WHH170 pKa = 8.37SGAEE174 pKa = 4.0AGASDD179 pKa = 5.26LEE181 pKa = 4.45TDD183 pKa = 3.38EE184 pKa = 6.15HH185 pKa = 6.75YY186 pKa = 11.41DD187 pKa = 3.57PMNEE191 pKa = 3.77
MM1 pKa = 7.43SPNAYY6 pKa = 9.05VHH8 pKa = 6.67WDD10 pKa = 3.36DD11 pKa = 4.75EE12 pKa = 4.31KK13 pKa = 11.31RR14 pKa = 11.84LLFGFQGEE22 pKa = 4.58AEE24 pKa = 4.17HH25 pKa = 6.41EE26 pKa = 4.34KK27 pKa = 10.72PVFTWTYY34 pKa = 8.67STFEE38 pKa = 4.09EE39 pKa = 4.9YY40 pKa = 10.95LNEE43 pKa = 4.17ACTPSMYY50 pKa = 10.81QFMANYY56 pKa = 9.27IIEE59 pKa = 4.93EE60 pKa = 3.92ISKK63 pKa = 11.06DD64 pKa = 3.57EE65 pKa = 4.34LYY67 pKa = 10.68EE68 pKa = 3.95LQGGDD73 pKa = 3.75YY74 pKa = 10.93CPGRR78 pKa = 11.84LEE80 pKa = 4.15EE81 pKa = 4.36EE82 pKa = 4.13AVDD85 pKa = 4.92AYY87 pKa = 11.16FDD89 pKa = 3.96LPINEE94 pKa = 4.75RR95 pKa = 11.84IEE97 pKa = 4.03LHH99 pKa = 5.51NQEE102 pKa = 5.13LLNLYY107 pKa = 10.29AEE109 pKa = 4.79RR110 pKa = 11.84DD111 pKa = 3.41RR112 pKa = 11.84ATGKK116 pKa = 9.72EE117 pKa = 3.68AAFIDD122 pKa = 3.93ACLDD126 pKa = 3.39DD127 pKa = 4.99HH128 pKa = 7.53CPFDD132 pKa = 4.3VDD134 pKa = 3.74SPIYY138 pKa = 10.65VEE140 pKa = 4.09YY141 pKa = 10.63CQWCRR146 pKa = 11.84KK147 pKa = 7.24QTEE150 pKa = 3.84KK151 pKa = 7.96WTKK154 pKa = 9.63IVARR158 pKa = 11.84LADD161 pKa = 4.87EE162 pKa = 3.95IHH164 pKa = 6.87EE165 pKa = 4.37EE166 pKa = 4.25EE167 pKa = 4.79DD168 pKa = 3.19WHH170 pKa = 8.37SGAEE174 pKa = 4.0AGASDD179 pKa = 5.26LEE181 pKa = 4.45TDD183 pKa = 3.38EE184 pKa = 6.15HH185 pKa = 6.75YY186 pKa = 11.41DD187 pKa = 3.57PMNEE191 pKa = 3.77
Molecular weight: 22.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0N7KVY5|A0A0N7KVY5_9PHYC Uncharacterized protein OS=Yellowstone lake phycodnavirus 3 OX=1586715 PE=4 SV=1
MM1 pKa = 6.04TTTRR5 pKa = 11.84IRR7 pKa = 11.84RR8 pKa = 11.84TTSPARR14 pKa = 11.84TAKK17 pKa = 8.95KK18 pKa = 7.55TASPARR24 pKa = 11.84TASPANKK31 pKa = 9.76RR32 pKa = 11.84PRR34 pKa = 11.84NAPNPWAPKK43 pKa = 7.98YY44 pKa = 8.69TWAPWGTSGVVHH56 pKa = 6.75DD57 pKa = 4.59NCYY60 pKa = 11.01DD61 pKa = 3.52YY62 pKa = 11.94AFGSFSNNRR71 pKa = 11.84TEE73 pKa = 3.96KK74 pKa = 10.39SVPGVRR80 pKa = 11.84AKK82 pKa = 10.95LNSNGMNFRR91 pKa = 11.84TCDD94 pKa = 3.77GIAKK98 pKa = 9.83RR99 pKa = 11.84VLADD103 pKa = 3.43NPGNVYY109 pKa = 10.57KK110 pKa = 10.42MKK112 pKa = 10.86SGDD115 pKa = 3.93EE116 pKa = 3.96KK117 pKa = 10.95PKK119 pKa = 10.16AGFYY123 pKa = 10.9KK124 pKa = 10.35VMCFVAPTNDD134 pKa = 3.4FGNSTGDD141 pKa = 3.19FHH143 pKa = 6.94WYY145 pKa = 10.31KK146 pKa = 10.28EE147 pKa = 3.99ISAIRR152 pKa = 11.84YY153 pKa = 7.43KK154 pKa = 10.2IRR156 pKa = 11.84RR157 pKa = 11.84ADD159 pKa = 3.79TIMGMAKK166 pKa = 10.07FFHH169 pKa = 5.89VTPKK173 pKa = 9.93TIRR176 pKa = 11.84DD177 pKa = 4.1AIAKK181 pKa = 9.37GRR183 pKa = 11.84ASANANNGRR192 pKa = 11.84VANKK196 pKa = 8.59NTDD199 pKa = 3.32LHH201 pKa = 6.51VLNSMSAASRR211 pKa = 11.84NKK213 pKa = 8.76MPVPVGRR220 pKa = 11.84VIDD223 pKa = 4.05FPVKK227 pKa = 10.15LWSHH231 pKa = 4.92KK232 pKa = 9.14TGWAGGPLIVDD243 pKa = 4.09ASGKK247 pKa = 9.06TITDD251 pKa = 3.08PRR253 pKa = 11.84RR254 pKa = 11.84CDD256 pKa = 3.12RR257 pKa = 11.84KK258 pKa = 8.82FTPGFHH264 pKa = 4.95YY265 pKa = 9.79TKK267 pKa = 10.11FCSAYY272 pKa = 8.73GVRR275 pKa = 11.84RR276 pKa = 11.84GFAKK280 pKa = 10.07TGNNANRR287 pKa = 11.84NN288 pKa = 3.73
MM1 pKa = 6.04TTTRR5 pKa = 11.84IRR7 pKa = 11.84RR8 pKa = 11.84TTSPARR14 pKa = 11.84TAKK17 pKa = 8.95KK18 pKa = 7.55TASPARR24 pKa = 11.84TASPANKK31 pKa = 9.76RR32 pKa = 11.84PRR34 pKa = 11.84NAPNPWAPKK43 pKa = 7.98YY44 pKa = 8.69TWAPWGTSGVVHH56 pKa = 6.75DD57 pKa = 4.59NCYY60 pKa = 11.01DD61 pKa = 3.52YY62 pKa = 11.94AFGSFSNNRR71 pKa = 11.84TEE73 pKa = 3.96KK74 pKa = 10.39SVPGVRR80 pKa = 11.84AKK82 pKa = 10.95LNSNGMNFRR91 pKa = 11.84TCDD94 pKa = 3.77GIAKK98 pKa = 9.83RR99 pKa = 11.84VLADD103 pKa = 3.43NPGNVYY109 pKa = 10.57KK110 pKa = 10.42MKK112 pKa = 10.86SGDD115 pKa = 3.93EE116 pKa = 3.96KK117 pKa = 10.95PKK119 pKa = 10.16AGFYY123 pKa = 10.9KK124 pKa = 10.35VMCFVAPTNDD134 pKa = 3.4FGNSTGDD141 pKa = 3.19FHH143 pKa = 6.94WYY145 pKa = 10.31KK146 pKa = 10.28EE147 pKa = 3.99ISAIRR152 pKa = 11.84YY153 pKa = 7.43KK154 pKa = 10.2IRR156 pKa = 11.84RR157 pKa = 11.84ADD159 pKa = 3.79TIMGMAKK166 pKa = 10.07FFHH169 pKa = 5.89VTPKK173 pKa = 9.93TIRR176 pKa = 11.84DD177 pKa = 4.1AIAKK181 pKa = 9.37GRR183 pKa = 11.84ASANANNGRR192 pKa = 11.84VANKK196 pKa = 8.59NTDD199 pKa = 3.32LHH201 pKa = 6.51VLNSMSAASRR211 pKa = 11.84NKK213 pKa = 8.76MPVPVGRR220 pKa = 11.84VIDD223 pKa = 4.05FPVKK227 pKa = 10.15LWSHH231 pKa = 4.92KK232 pKa = 9.14TGWAGGPLIVDD243 pKa = 4.09ASGKK247 pKa = 9.06TITDD251 pKa = 3.08PRR253 pKa = 11.84RR254 pKa = 11.84CDD256 pKa = 3.12RR257 pKa = 11.84KK258 pKa = 8.82FTPGFHH264 pKa = 4.95YY265 pKa = 9.79TKK267 pKa = 10.11FCSAYY272 pKa = 8.73GVRR275 pKa = 11.84RR276 pKa = 11.84GFAKK280 pKa = 10.07TGNNANRR287 pKa = 11.84NN288 pKa = 3.73
Molecular weight: 31.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
53188 |
53 |
1432 |
225.4 |
24.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.634 ± 0.313 | 1.542 ± 0.122 |
4.715 ± 0.26 | 5.076 ± 0.294 |
4.172 ± 0.117 | 7.165 ± 0.227 |
1.688 ± 0.122 | 5.334 ± 0.11 |
6.067 ± 0.334 | 8.062 ± 0.166 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.76 ± 0.119 | 5.443 ± 0.32 |
5.356 ± 0.14 | 3.495 ± 0.112 |
5.721 ± 0.35 | 6.515 ± 0.287 |
6.618 ± 0.448 | 7.069 ± 0.204 |
1.324 ± 0.068 | 3.243 ± 0.101 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
