
Opium poppy mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Calvusvirinae; Umbravirus
Average proteome isoelectric point is 7.57
Get precalculated fractions of proteins
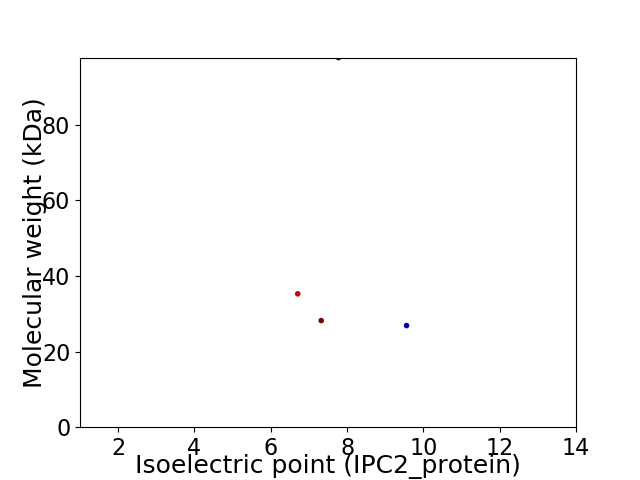
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
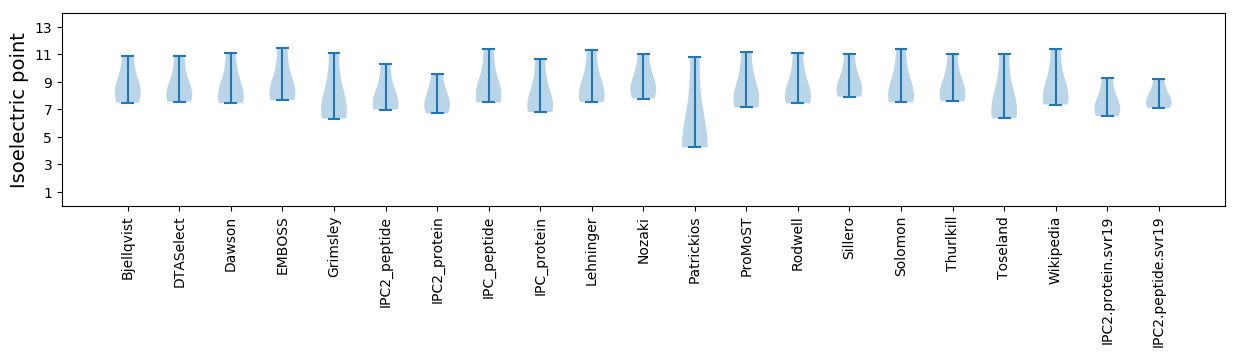
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G0KUS6|G0KUS6_9TOMB Putative phloem RNA movement protein OS=Opium poppy mosaic virus OX=473784 PE=4 SV=2
MM1 pKa = 7.88DD2 pKa = 4.05FVDD5 pKa = 6.31RR6 pKa = 11.84ITRR9 pKa = 11.84WLSGDD14 pKa = 4.47FKK16 pKa = 10.69PSAYY20 pKa = 9.23IKK22 pKa = 10.56SRR24 pKa = 11.84NEE26 pKa = 3.56CVVAYY31 pKa = 10.15GEE33 pKa = 4.13EE34 pKa = 4.19WATLVTQSRR43 pKa = 11.84MQRR46 pKa = 11.84EE47 pKa = 4.24ASRR50 pKa = 11.84QYY52 pKa = 11.54ADD54 pKa = 3.56WYY56 pKa = 10.84NSASPTTCTMPPPTDD71 pKa = 3.32AEE73 pKa = 4.6LIEE76 pKa = 4.12EE77 pKa = 4.16ARR79 pKa = 11.84VGKK82 pKa = 10.39LKK84 pKa = 10.95AGLDD88 pKa = 3.65QPKK91 pKa = 9.49KK92 pKa = 7.79QQEE95 pKa = 3.95AASINLGGIKK105 pKa = 9.19TPPPMSEE112 pKa = 4.06EE113 pKa = 3.9QSTLLDD119 pKa = 3.84LEE121 pKa = 4.79ALPSYY126 pKa = 10.33DD127 pKa = 3.78QIVAVGKK134 pKa = 9.96LRR136 pKa = 11.84RR137 pKa = 11.84LQLQSACPQYY147 pKa = 10.68TDD149 pKa = 3.76EE150 pKa = 5.38EE151 pKa = 4.35RR152 pKa = 11.84ASWAIIPYY160 pKa = 8.74TGGPRR165 pKa = 11.84VIRR168 pKa = 11.84EE169 pKa = 4.14DD170 pKa = 3.49EE171 pKa = 4.22VLPRR175 pKa = 11.84APSVVLLNDD184 pKa = 3.83APRR187 pKa = 11.84SMSKK191 pKa = 9.62IEE193 pKa = 4.19EE194 pKa = 4.15LVKK197 pKa = 10.99ACFATIDD204 pKa = 4.36KK205 pKa = 9.0VAKK208 pKa = 8.0WRR210 pKa = 11.84KK211 pKa = 8.43VDD213 pKa = 4.02YY214 pKa = 10.8IDD216 pKa = 4.7FSSKK220 pKa = 10.46AAQTPPSKK228 pKa = 11.03YY229 pKa = 9.9KK230 pKa = 10.22LQEE233 pKa = 4.17PGEE236 pKa = 4.33VMKK239 pKa = 10.91GMNAQSVAMEE249 pKa = 3.93LRR251 pKa = 11.84CRR253 pKa = 11.84YY254 pKa = 8.86GVQPCTAANLQLGNRR269 pKa = 11.84VARR272 pKa = 11.84EE273 pKa = 3.55ILDD276 pKa = 3.83RR277 pKa = 11.84QCQATRR283 pKa = 11.84EE284 pKa = 3.87QVYY287 pKa = 10.04YY288 pKa = 10.02YY289 pKa = 11.0GHH291 pKa = 7.86LATTMWFQPTLVDD304 pKa = 3.84LALRR308 pKa = 11.84AGAKK312 pKa = 10.32DD313 pKa = 3.36FF314 pKa = 4.45
MM1 pKa = 7.88DD2 pKa = 4.05FVDD5 pKa = 6.31RR6 pKa = 11.84ITRR9 pKa = 11.84WLSGDD14 pKa = 4.47FKK16 pKa = 10.69PSAYY20 pKa = 9.23IKK22 pKa = 10.56SRR24 pKa = 11.84NEE26 pKa = 3.56CVVAYY31 pKa = 10.15GEE33 pKa = 4.13EE34 pKa = 4.19WATLVTQSRR43 pKa = 11.84MQRR46 pKa = 11.84EE47 pKa = 4.24ASRR50 pKa = 11.84QYY52 pKa = 11.54ADD54 pKa = 3.56WYY56 pKa = 10.84NSASPTTCTMPPPTDD71 pKa = 3.32AEE73 pKa = 4.6LIEE76 pKa = 4.12EE77 pKa = 4.16ARR79 pKa = 11.84VGKK82 pKa = 10.39LKK84 pKa = 10.95AGLDD88 pKa = 3.65QPKK91 pKa = 9.49KK92 pKa = 7.79QQEE95 pKa = 3.95AASINLGGIKK105 pKa = 9.19TPPPMSEE112 pKa = 4.06EE113 pKa = 3.9QSTLLDD119 pKa = 3.84LEE121 pKa = 4.79ALPSYY126 pKa = 10.33DD127 pKa = 3.78QIVAVGKK134 pKa = 9.96LRR136 pKa = 11.84RR137 pKa = 11.84LQLQSACPQYY147 pKa = 10.68TDD149 pKa = 3.76EE150 pKa = 5.38EE151 pKa = 4.35RR152 pKa = 11.84ASWAIIPYY160 pKa = 8.74TGGPRR165 pKa = 11.84VIRR168 pKa = 11.84EE169 pKa = 4.14DD170 pKa = 3.49EE171 pKa = 4.22VLPRR175 pKa = 11.84APSVVLLNDD184 pKa = 3.83APRR187 pKa = 11.84SMSKK191 pKa = 9.62IEE193 pKa = 4.19EE194 pKa = 4.15LVKK197 pKa = 10.99ACFATIDD204 pKa = 4.36KK205 pKa = 9.0VAKK208 pKa = 8.0WRR210 pKa = 11.84KK211 pKa = 8.43VDD213 pKa = 4.02YY214 pKa = 10.8IDD216 pKa = 4.7FSSKK220 pKa = 10.46AAQTPPSKK228 pKa = 11.03YY229 pKa = 9.9KK230 pKa = 10.22LQEE233 pKa = 4.17PGEE236 pKa = 4.33VMKK239 pKa = 10.91GMNAQSVAMEE249 pKa = 3.93LRR251 pKa = 11.84CRR253 pKa = 11.84YY254 pKa = 8.86GVQPCTAANLQLGNRR269 pKa = 11.84VARR272 pKa = 11.84EE273 pKa = 3.55ILDD276 pKa = 3.83RR277 pKa = 11.84QCQATRR283 pKa = 11.84EE284 pKa = 3.87QVYY287 pKa = 10.04YY288 pKa = 10.02YY289 pKa = 11.0GHH291 pKa = 7.86LATTMWFQPTLVDD304 pKa = 3.84LALRR308 pKa = 11.84AGAKK312 pKa = 10.32DD313 pKa = 3.36FF314 pKa = 4.45
Molecular weight: 35.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G0KUS7|G0KUS7_9TOMB Putative cell-to-cell movement protein OS=Opium poppy mosaic virus OX=473784 PE=4 SV=2
MM1 pKa = 7.73ASLPNVSSSSKK12 pKa = 10.75SRR14 pKa = 11.84MQGRR18 pKa = 11.84TTQRR22 pKa = 11.84AIRR25 pKa = 11.84RR26 pKa = 11.84GDD28 pKa = 3.32GQRR31 pKa = 11.84ARR33 pKa = 11.84RR34 pKa = 11.84NRR36 pKa = 11.84PGHH39 pKa = 5.95TDD41 pKa = 3.46PPPSRR46 pKa = 11.84AEE48 pKa = 3.72ACRR51 pKa = 11.84NPAPAPKK58 pKa = 10.42NPGKK62 pKa = 10.27NRR64 pKa = 11.84GSLEE68 pKa = 3.99EE69 pKa = 4.2TPHH72 pKa = 5.77HH73 pKa = 6.51KK74 pKa = 9.97KK75 pKa = 9.9HH76 pKa = 6.57RR77 pKa = 11.84GPVVPRR83 pKa = 11.84EE84 pKa = 4.25NRR86 pKa = 11.84GSVHH90 pKa = 4.96PTRR93 pKa = 11.84PRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84GDD102 pKa = 3.08HH103 pKa = 5.57MGSRR107 pKa = 11.84QHH109 pKa = 6.22ATTPEE114 pKa = 4.17QCWGACVLSTEE125 pKa = 4.2RR126 pKa = 11.84RR127 pKa = 11.84AEE129 pKa = 3.82IDD131 pKa = 3.54GLLSPLLHH139 pKa = 6.16TALMQGWGDD148 pKa = 3.84PEE150 pKa = 4.09VLVYY154 pKa = 9.8SIRR157 pKa = 11.84VLRR160 pKa = 11.84GEE162 pKa = 3.8LRR164 pKa = 11.84QWRR167 pKa = 11.84KK168 pKa = 8.73PVQSLHH174 pKa = 6.07NVATEE179 pKa = 3.85DD180 pKa = 3.65RR181 pKa = 11.84GNSAQLSAPIATEE194 pKa = 4.07PPNMQAPSKK203 pKa = 8.99EE204 pKa = 3.9QPAVDD209 pKa = 4.76GSPKK213 pKa = 9.86ICNLGCNIDD222 pKa = 4.12KK223 pKa = 11.0VCGHH227 pKa = 7.29PKK229 pKa = 9.87PGEE232 pKa = 4.28GNSGSTTQAGNTWW245 pKa = 3.06
MM1 pKa = 7.73ASLPNVSSSSKK12 pKa = 10.75SRR14 pKa = 11.84MQGRR18 pKa = 11.84TTQRR22 pKa = 11.84AIRR25 pKa = 11.84RR26 pKa = 11.84GDD28 pKa = 3.32GQRR31 pKa = 11.84ARR33 pKa = 11.84RR34 pKa = 11.84NRR36 pKa = 11.84PGHH39 pKa = 5.95TDD41 pKa = 3.46PPPSRR46 pKa = 11.84AEE48 pKa = 3.72ACRR51 pKa = 11.84NPAPAPKK58 pKa = 10.42NPGKK62 pKa = 10.27NRR64 pKa = 11.84GSLEE68 pKa = 3.99EE69 pKa = 4.2TPHH72 pKa = 5.77HH73 pKa = 6.51KK74 pKa = 9.97KK75 pKa = 9.9HH76 pKa = 6.57RR77 pKa = 11.84GPVVPRR83 pKa = 11.84EE84 pKa = 4.25NRR86 pKa = 11.84GSVHH90 pKa = 4.96PTRR93 pKa = 11.84PRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84GDD102 pKa = 3.08HH103 pKa = 5.57MGSRR107 pKa = 11.84QHH109 pKa = 6.22ATTPEE114 pKa = 4.17QCWGACVLSTEE125 pKa = 4.2RR126 pKa = 11.84RR127 pKa = 11.84AEE129 pKa = 3.82IDD131 pKa = 3.54GLLSPLLHH139 pKa = 6.16TALMQGWGDD148 pKa = 3.84PEE150 pKa = 4.09VLVYY154 pKa = 9.8SIRR157 pKa = 11.84VLRR160 pKa = 11.84GEE162 pKa = 3.8LRR164 pKa = 11.84QWRR167 pKa = 11.84KK168 pKa = 8.73PVQSLHH174 pKa = 6.07NVATEE179 pKa = 3.85DD180 pKa = 3.65RR181 pKa = 11.84GNSAQLSAPIATEE194 pKa = 4.07PPNMQAPSKK203 pKa = 8.99EE204 pKa = 3.9QPAVDD209 pKa = 4.76GSPKK213 pKa = 9.86ICNLGCNIDD222 pKa = 4.12KK223 pKa = 11.0VCGHH227 pKa = 7.29PKK229 pKa = 9.87PGEE232 pKa = 4.28GNSGSTTQAGNTWW245 pKa = 3.06
Molecular weight: 26.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1681 |
245 |
865 |
420.3 |
47.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.626 ± 0.457 | 2.32 ± 0.089 |
4.819 ± 0.75 | 6.068 ± 0.333 |
2.558 ± 0.638 | 6.246 ± 0.603 |
2.142 ± 0.483 | 4.521 ± 0.449 |
5.473 ± 0.558 | 8.15 ± 0.593 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.677 ± 0.25 | 3.629 ± 0.427 |
7.02 ± 0.905 | 4.521 ± 0.57 |
7.436 ± 0.885 | 6.425 ± 0.455 |
5.532 ± 0.134 | 6.901 ± 0.438 |
1.725 ± 0.219 | 3.212 ± 0.545 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
