
Murine feces-associated gemycircularvirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; unclassified Gemycircularvirus
Average proteome isoelectric point is 8.9
Get precalculated fractions of proteins
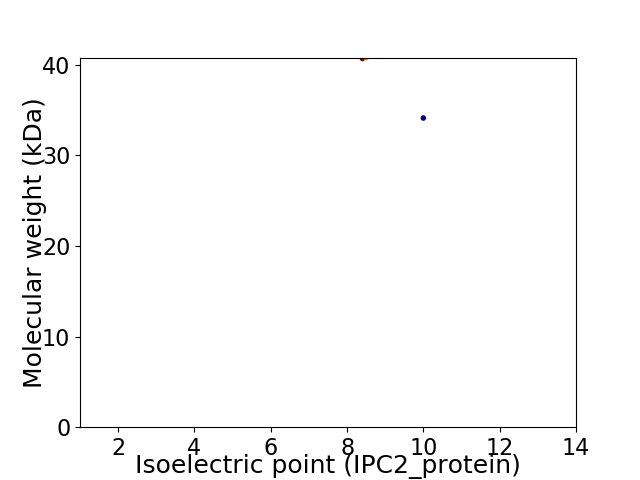
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S0SZ51|A0A2S0SZ51_9VIRU Cap OS=Murine feces-associated gemycircularvirus 2 OX=2171385 PE=4 SV=1
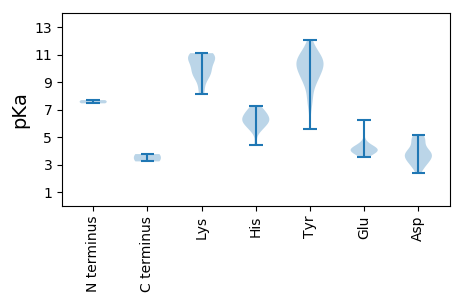
MM1 pKa = 7.45SNDD4 pKa = 2.79SSTRR8 pKa = 11.84EE9 pKa = 4.04LLSLTVSRR17 pKa = 11.84RR18 pKa = 11.84ILEE21 pKa = 4.03ILDD24 pKa = 3.8AVGSDD29 pKa = 3.51ASITAMEE36 pKa = 4.16ASITTFTLVSTRR48 pKa = 11.84LQLSTRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 9.07RR57 pKa = 11.84SIISEE62 pKa = 3.94LTEE65 pKa = 3.58ILNQYY70 pKa = 8.95EE71 pKa = 3.99EE72 pKa = 4.4HH73 pKa = 5.99QRR75 pKa = 11.84SLLTMQARR83 pKa = 11.84RR84 pKa = 11.84EE85 pKa = 4.12MYY87 pKa = 10.24DD88 pKa = 2.64WTTANPHH95 pKa = 6.03DD96 pKa = 4.57HH97 pKa = 6.87LLNVASMIQINGQISSAAQIKK118 pKa = 9.59NHH120 pKa = 6.19FCTLYY125 pKa = 10.23VRR127 pKa = 11.84RR128 pKa = 11.84LQEE131 pKa = 3.69IGCYY135 pKa = 9.19PIRR138 pKa = 11.84GSSSMPTYY146 pKa = 9.64TIPTRR151 pKa = 11.84SPNINHH157 pKa = 6.72RR158 pKa = 11.84PSLHH162 pKa = 6.58RR163 pKa = 11.84MNATLSSSNGSNRR176 pKa = 11.84HH177 pKa = 4.41TLEE180 pKa = 3.77SHH182 pKa = 5.56EE183 pKa = 4.26MRR185 pKa = 11.84GKK187 pKa = 10.87LSLLAAGPAPGCGNRR202 pKa = 11.84RR203 pKa = 11.84GGCAPTPLRR212 pKa = 11.84TEE214 pKa = 4.16WNWANWRR221 pKa = 11.84RR222 pKa = 11.84VKK224 pKa = 10.84SLILWGGTRR233 pKa = 11.84TGKK236 pKa = 8.13TLYY239 pKa = 10.48ARR241 pKa = 11.84SLGHH245 pKa = 6.0HH246 pKa = 7.26AYY248 pKa = 11.04YY249 pKa = 10.88NLQFNMDD256 pKa = 4.81DD257 pKa = 3.87FSNDD261 pKa = 2.4CKK263 pKa = 11.1YY264 pKa = 11.29AVFDD268 pKa = 5.17DD269 pKa = 3.78IQGGFEE275 pKa = 3.92YY276 pKa = 9.65WHH278 pKa = 6.92SYY280 pKa = 9.54KK281 pKa = 10.77GWLGAQKK288 pKa = 10.48QFVITDD294 pKa = 3.92KK295 pKa = 10.7YY296 pKa = 9.09RR297 pKa = 11.84KK298 pKa = 9.5KK299 pKa = 10.11RR300 pKa = 11.84TIHH303 pKa = 5.65WGKK306 pKa = 9.61PSIMCTNDD314 pKa = 3.51DD315 pKa = 4.26PFSMRR320 pKa = 11.84NVDD323 pKa = 4.88YY324 pKa = 10.98DD325 pKa = 3.24WLMNNCYY332 pKa = 9.79IINVVDD338 pKa = 4.89PICIVSSLPSSSDD351 pKa = 3.34NQSEE355 pKa = 4.35QII357 pKa = 3.76
MM1 pKa = 7.45SNDD4 pKa = 2.79SSTRR8 pKa = 11.84EE9 pKa = 4.04LLSLTVSRR17 pKa = 11.84RR18 pKa = 11.84ILEE21 pKa = 4.03ILDD24 pKa = 3.8AVGSDD29 pKa = 3.51ASITAMEE36 pKa = 4.16ASITTFTLVSTRR48 pKa = 11.84LQLSTRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 9.07RR57 pKa = 11.84SIISEE62 pKa = 3.94LTEE65 pKa = 3.58ILNQYY70 pKa = 8.95EE71 pKa = 3.99EE72 pKa = 4.4HH73 pKa = 5.99QRR75 pKa = 11.84SLLTMQARR83 pKa = 11.84RR84 pKa = 11.84EE85 pKa = 4.12MYY87 pKa = 10.24DD88 pKa = 2.64WTTANPHH95 pKa = 6.03DD96 pKa = 4.57HH97 pKa = 6.87LLNVASMIQINGQISSAAQIKK118 pKa = 9.59NHH120 pKa = 6.19FCTLYY125 pKa = 10.23VRR127 pKa = 11.84RR128 pKa = 11.84LQEE131 pKa = 3.69IGCYY135 pKa = 9.19PIRR138 pKa = 11.84GSSSMPTYY146 pKa = 9.64TIPTRR151 pKa = 11.84SPNINHH157 pKa = 6.72RR158 pKa = 11.84PSLHH162 pKa = 6.58RR163 pKa = 11.84MNATLSSSNGSNRR176 pKa = 11.84HH177 pKa = 4.41TLEE180 pKa = 3.77SHH182 pKa = 5.56EE183 pKa = 4.26MRR185 pKa = 11.84GKK187 pKa = 10.87LSLLAAGPAPGCGNRR202 pKa = 11.84RR203 pKa = 11.84GGCAPTPLRR212 pKa = 11.84TEE214 pKa = 4.16WNWANWRR221 pKa = 11.84RR222 pKa = 11.84VKK224 pKa = 10.84SLILWGGTRR233 pKa = 11.84TGKK236 pKa = 8.13TLYY239 pKa = 10.48ARR241 pKa = 11.84SLGHH245 pKa = 6.0HH246 pKa = 7.26AYY248 pKa = 11.04YY249 pKa = 10.88NLQFNMDD256 pKa = 4.81DD257 pKa = 3.87FSNDD261 pKa = 2.4CKK263 pKa = 11.1YY264 pKa = 11.29AVFDD268 pKa = 5.17DD269 pKa = 3.78IQGGFEE275 pKa = 3.92YY276 pKa = 9.65WHH278 pKa = 6.92SYY280 pKa = 9.54KK281 pKa = 10.77GWLGAQKK288 pKa = 10.48QFVITDD294 pKa = 3.92KK295 pKa = 10.7YY296 pKa = 9.09RR297 pKa = 11.84KK298 pKa = 9.5KK299 pKa = 10.11RR300 pKa = 11.84TIHH303 pKa = 5.65WGKK306 pKa = 9.61PSIMCTNDD314 pKa = 3.51DD315 pKa = 4.26PFSMRR320 pKa = 11.84NVDD323 pKa = 4.88YY324 pKa = 10.98DD325 pKa = 3.24WLMNNCYY332 pKa = 9.79IINVVDD338 pKa = 4.89PICIVSSLPSSSDD351 pKa = 3.34NQSEE355 pKa = 4.35QII357 pKa = 3.76
Molecular weight: 40.7 kDa
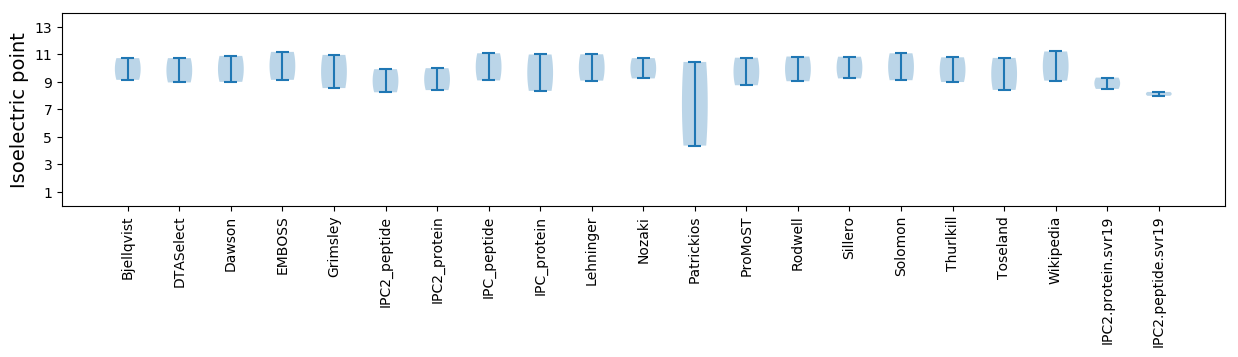
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S0SZ51|A0A2S0SZ51_9VIRU Cap OS=Murine feces-associated gemycircularvirus 2 OX=2171385 PE=4 SV=1
MM1 pKa = 7.69ALRR4 pKa = 11.84SRR6 pKa = 11.84FYY8 pKa = 10.18RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84MGRR15 pKa = 11.84SYY17 pKa = 10.62RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84PVRR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 8.9SRR27 pKa = 11.84RR28 pKa = 11.84GRR30 pKa = 11.84SGYY33 pKa = 10.52GRR35 pKa = 11.84GGYY38 pKa = 7.78NRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84YY43 pKa = 9.23RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SWISPYY54 pKa = 10.45DD55 pKa = 3.53KK56 pKa = 10.94KK57 pKa = 11.1RR58 pKa = 11.84DD59 pKa = 3.9VVKK62 pKa = 10.51GAHH65 pKa = 6.16DD66 pKa = 3.68TNDD69 pKa = 3.08NSFAPIGAPNTFFGFCPTYY88 pKa = 10.07MPNAVAGDD96 pKa = 3.98TGHH99 pKa = 6.19SHH101 pKa = 5.51QKK103 pKa = 9.93VRR105 pKa = 11.84MSKK108 pKa = 9.51NVGFTGYY115 pKa = 7.79QEE117 pKa = 4.12RR118 pKa = 11.84VHH120 pKa = 6.38IATAEE125 pKa = 3.98SLIWRR130 pKa = 11.84RR131 pKa = 11.84IVIWSYY137 pKa = 11.43NRR139 pKa = 11.84LNNTAGPTKK148 pKa = 9.82TDD150 pKa = 3.12SDD152 pKa = 4.11GVSYY156 pKa = 7.39TTRR159 pKa = 11.84QITPLQLTEE168 pKa = 3.85EE169 pKa = 4.21VRR171 pKa = 11.84RR172 pKa = 11.84FLFRR176 pKa = 11.84GTQGVDD182 pKa = 3.16YY183 pKa = 10.72TEE185 pKa = 4.36NTLHH189 pKa = 6.48EE190 pKa = 4.74APLNADD196 pKa = 3.86HH197 pKa = 7.03FVVAYY202 pKa = 10.34DD203 pKa = 3.6RR204 pKa = 11.84TRR206 pKa = 11.84VLQPRR211 pKa = 11.84RR212 pKa = 11.84PTGDD216 pKa = 2.99EE217 pKa = 3.82YY218 pKa = 12.03GHH220 pKa = 6.44IFNMKK225 pKa = 8.4FWNPGGRR232 pKa = 11.84IIYY235 pKa = 10.4SDD237 pKa = 4.94DD238 pKa = 3.49EE239 pKa = 6.21DD240 pKa = 5.15GLTKK244 pKa = 10.78GPLVNGWSSLGRR256 pKa = 11.84ASKK259 pKa = 11.08GNMYY263 pKa = 10.41IFDD266 pKa = 4.53LFSTGFSLTSNTPSIARR283 pKa = 11.84FAPTGCRR290 pKa = 11.84YY291 pKa = 5.61WTEE294 pKa = 4.05GG295 pKa = 3.23
MM1 pKa = 7.69ALRR4 pKa = 11.84SRR6 pKa = 11.84FYY8 pKa = 10.18RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84MGRR15 pKa = 11.84SYY17 pKa = 10.62RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84PVRR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 8.9SRR27 pKa = 11.84RR28 pKa = 11.84GRR30 pKa = 11.84SGYY33 pKa = 10.52GRR35 pKa = 11.84GGYY38 pKa = 7.78NRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84YY43 pKa = 9.23RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SWISPYY54 pKa = 10.45DD55 pKa = 3.53KK56 pKa = 10.94KK57 pKa = 11.1RR58 pKa = 11.84DD59 pKa = 3.9VVKK62 pKa = 10.51GAHH65 pKa = 6.16DD66 pKa = 3.68TNDD69 pKa = 3.08NSFAPIGAPNTFFGFCPTYY88 pKa = 10.07MPNAVAGDD96 pKa = 3.98TGHH99 pKa = 6.19SHH101 pKa = 5.51QKK103 pKa = 9.93VRR105 pKa = 11.84MSKK108 pKa = 9.51NVGFTGYY115 pKa = 7.79QEE117 pKa = 4.12RR118 pKa = 11.84VHH120 pKa = 6.38IATAEE125 pKa = 3.98SLIWRR130 pKa = 11.84RR131 pKa = 11.84IVIWSYY137 pKa = 11.43NRR139 pKa = 11.84LNNTAGPTKK148 pKa = 9.82TDD150 pKa = 3.12SDD152 pKa = 4.11GVSYY156 pKa = 7.39TTRR159 pKa = 11.84QITPLQLTEE168 pKa = 3.85EE169 pKa = 4.21VRR171 pKa = 11.84RR172 pKa = 11.84FLFRR176 pKa = 11.84GTQGVDD182 pKa = 3.16YY183 pKa = 10.72TEE185 pKa = 4.36NTLHH189 pKa = 6.48EE190 pKa = 4.74APLNADD196 pKa = 3.86HH197 pKa = 7.03FVVAYY202 pKa = 10.34DD203 pKa = 3.6RR204 pKa = 11.84TRR206 pKa = 11.84VLQPRR211 pKa = 11.84RR212 pKa = 11.84PTGDD216 pKa = 2.99EE217 pKa = 3.82YY218 pKa = 12.03GHH220 pKa = 6.44IFNMKK225 pKa = 8.4FWNPGGRR232 pKa = 11.84IIYY235 pKa = 10.4SDD237 pKa = 4.94DD238 pKa = 3.49EE239 pKa = 6.21DD240 pKa = 5.15GLTKK244 pKa = 10.78GPLVNGWSSLGRR256 pKa = 11.84ASKK259 pKa = 11.08GNMYY263 pKa = 10.41IFDD266 pKa = 4.53LFSTGFSLTSNTPSIARR283 pKa = 11.84FAPTGCRR290 pKa = 11.84YY291 pKa = 5.61WTEE294 pKa = 4.05GG295 pKa = 3.23
Molecular weight: 34.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
652 |
295 |
357 |
326.0 |
37.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.215 ± 0.078 | 1.534 ± 0.514 |
4.908 ± 0.106 | 3.528 ± 0.286 |
3.528 ± 0.935 | 7.515 ± 1.39 |
2.914 ± 0.325 | 5.675 ± 0.965 |
3.221 ± 0.102 | 6.902 ± 1.294 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.761 ± 0.436 | 5.982 ± 0.335 |
4.448 ± 0.382 | 3.067 ± 0.621 |
10.736 ± 2.102 | 9.202 ± 1.251 |
7.669 ± 0.28 | 4.141 ± 0.567 |
2.301 ± 0.16 | 4.755 ± 0.605 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
