
Hubei myriapoda virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
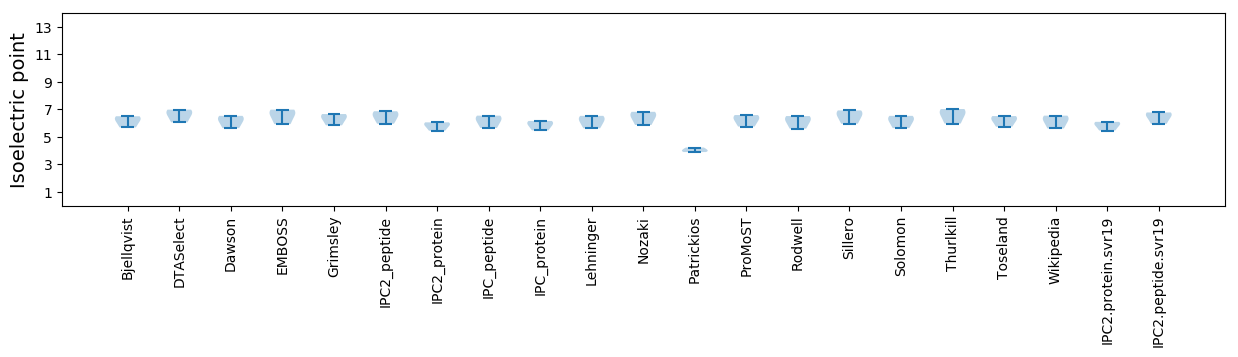
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
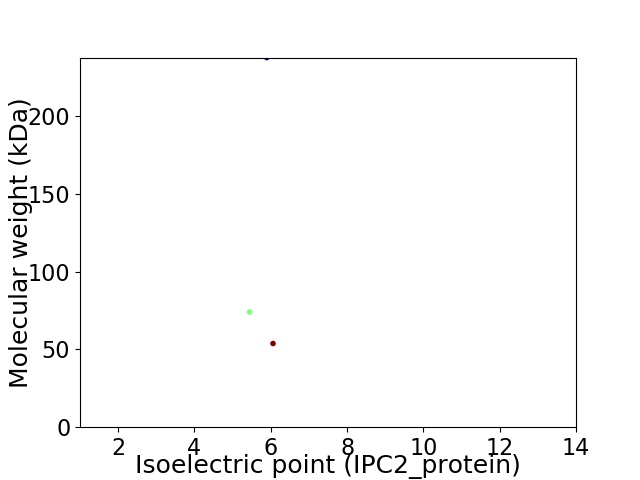
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLZ2|A0A1L3KLZ2_9VIRU Uncharacterized protein OS=Hubei myriapoda virus 2 OX=1922931 PE=4 SV=1
MM1 pKa = 7.84ADD3 pKa = 2.96GRR5 pKa = 11.84APAVPDD11 pKa = 3.51AEE13 pKa = 4.06AAEE16 pKa = 4.22TGEE19 pKa = 4.47IISTQTVPPTVPASRR34 pKa = 11.84VASGPPPSEE43 pKa = 3.83IVQHH47 pKa = 6.05TGNANLIDD55 pKa = 4.37PNILNQNILSSTITWSVNQPQNSVLGTFYY84 pKa = 11.01VHH86 pKa = 7.08PSYY89 pKa = 10.97HH90 pKa = 6.87LYY92 pKa = 10.8LDD94 pKa = 3.73YY95 pKa = 11.12LSRR98 pKa = 11.84IYY100 pKa = 8.57YY101 pKa = 7.26TWSGSLQYY109 pKa = 10.23SVCVLGTGFHH119 pKa = 6.91AGKK122 pKa = 10.29IMLVRR127 pKa = 11.84APPGYY132 pKa = 10.79DD133 pKa = 3.68LDD135 pKa = 3.88SLSNIEE141 pKa = 4.23EE142 pKa = 4.2ATGLEE147 pKa = 4.22WMALDD152 pKa = 4.29PKK154 pKa = 10.86SIIPAEE160 pKa = 4.35LGLNDD165 pKa = 3.29YY166 pKa = 10.87RR167 pKa = 11.84GVNFHH172 pKa = 6.04MTSIPADD179 pKa = 3.62SRR181 pKa = 11.84VTAAATKK188 pKa = 9.81TFACPEE194 pKa = 4.39GGWGRR199 pKa = 11.84LKK201 pKa = 10.61HH202 pKa = 6.88DD203 pKa = 4.18PDD205 pKa = 3.71KK206 pKa = 11.44VRR208 pKa = 11.84SAVRR212 pKa = 11.84AGQDD216 pKa = 3.1VSSFFDD222 pKa = 4.03TIRR225 pKa = 11.84HH226 pKa = 5.44GGILQLRR233 pKa = 11.84VMGEE237 pKa = 4.37LKK239 pKa = 10.16TSSTGNMQIQILVFVKK255 pKa = 10.26PGPDD259 pKa = 3.4FQFSTLIPPTVKK271 pKa = 10.51VGPTPSQVPAVIASLLSSAVAVRR294 pKa = 11.84RR295 pKa = 11.84CSATLSPVRR304 pKa = 11.84TLLVVSASVTPNKK317 pKa = 9.26MWFYY321 pKa = 11.44GCASSQGDD329 pKa = 3.25WCHH332 pKa = 5.16NTRR335 pKa = 11.84SYY337 pKa = 8.6VTKK340 pKa = 10.64DD341 pKa = 3.27PIITHH346 pKa = 5.63SPEE349 pKa = 3.34ISLLRR354 pKa = 11.84FPTVAKK360 pKa = 10.48NDD362 pKa = 3.49FSGYY366 pKa = 8.7VCDD369 pKa = 4.52VRR371 pKa = 11.84GATGNEE377 pKa = 4.36PIAQIAPKK385 pKa = 10.26GDD387 pKa = 3.32LGKK390 pKa = 10.79VVDD393 pKa = 4.82PFDD396 pKa = 5.14CNVLILEE403 pKa = 4.52NVVEE407 pKa = 4.27ANEE410 pKa = 4.14IRR412 pKa = 11.84GTEE415 pKa = 3.83YY416 pKa = 10.85DD417 pKa = 3.85APDD420 pKa = 3.8SVWIAEE426 pKa = 4.4EE427 pKa = 3.93NLSADD432 pKa = 3.78RR433 pKa = 11.84SEE435 pKa = 4.59VATVVHH441 pKa = 6.18PVGMWEE447 pKa = 5.05LPDD450 pKa = 3.68QEE452 pKa = 5.84LVFADD457 pKa = 3.67SAFMKK462 pKa = 9.09DD463 pKa = 2.64TWHH466 pKa = 6.72RR467 pKa = 11.84TLKK470 pKa = 10.19WPVNSVIDD478 pKa = 4.09RR479 pKa = 11.84EE480 pKa = 4.52SMTAKK485 pKa = 10.53LNLNFMPANAPVCWRR500 pKa = 11.84AIRR503 pKa = 11.84PVALDD508 pKa = 3.46LFIKK512 pKa = 10.56KK513 pKa = 9.65EE514 pKa = 3.9YY515 pKa = 10.87LNLKK519 pKa = 8.95PATKK523 pKa = 9.99EE524 pKa = 4.18SVFLFCPIKK533 pKa = 10.68PGTLDD538 pKa = 3.3AASIEE543 pKa = 4.23TGKK546 pKa = 11.01YY547 pKa = 10.16GLPTYY552 pKa = 9.45QTDD555 pKa = 3.47DD556 pKa = 3.2VSRR559 pKa = 11.84YY560 pKa = 8.85LAYY563 pKa = 10.63GNAQGLLTSEE573 pKa = 4.44DD574 pKa = 3.39ALLFEE579 pKa = 5.79LYY581 pKa = 10.1SVPLGTPVRR590 pKa = 11.84YY591 pKa = 10.0LKK593 pKa = 9.95MYY595 pKa = 9.11SSGMITTNASDD606 pKa = 3.36TTLILDD612 pKa = 4.04KK613 pKa = 11.25SDD615 pKa = 4.5DD616 pKa = 3.6YY617 pKa = 11.89DD618 pKa = 3.46VRR620 pKa = 11.84FVQKK624 pKa = 9.94IAASAAIPALPTHH637 pKa = 5.91STWASQVHH645 pKa = 5.92IANVVGKK652 pKa = 10.18RR653 pKa = 11.84EE654 pKa = 3.78NAMLRR659 pKa = 11.84EE660 pKa = 4.33SVTALDD666 pKa = 4.49RR667 pKa = 11.84KK668 pKa = 10.24IEE670 pKa = 4.08LLTSLLRR677 pKa = 11.84KK678 pKa = 9.53
MM1 pKa = 7.84ADD3 pKa = 2.96GRR5 pKa = 11.84APAVPDD11 pKa = 3.51AEE13 pKa = 4.06AAEE16 pKa = 4.22TGEE19 pKa = 4.47IISTQTVPPTVPASRR34 pKa = 11.84VASGPPPSEE43 pKa = 3.83IVQHH47 pKa = 6.05TGNANLIDD55 pKa = 4.37PNILNQNILSSTITWSVNQPQNSVLGTFYY84 pKa = 11.01VHH86 pKa = 7.08PSYY89 pKa = 10.97HH90 pKa = 6.87LYY92 pKa = 10.8LDD94 pKa = 3.73YY95 pKa = 11.12LSRR98 pKa = 11.84IYY100 pKa = 8.57YY101 pKa = 7.26TWSGSLQYY109 pKa = 10.23SVCVLGTGFHH119 pKa = 6.91AGKK122 pKa = 10.29IMLVRR127 pKa = 11.84APPGYY132 pKa = 10.79DD133 pKa = 3.68LDD135 pKa = 3.88SLSNIEE141 pKa = 4.23EE142 pKa = 4.2ATGLEE147 pKa = 4.22WMALDD152 pKa = 4.29PKK154 pKa = 10.86SIIPAEE160 pKa = 4.35LGLNDD165 pKa = 3.29YY166 pKa = 10.87RR167 pKa = 11.84GVNFHH172 pKa = 6.04MTSIPADD179 pKa = 3.62SRR181 pKa = 11.84VTAAATKK188 pKa = 9.81TFACPEE194 pKa = 4.39GGWGRR199 pKa = 11.84LKK201 pKa = 10.61HH202 pKa = 6.88DD203 pKa = 4.18PDD205 pKa = 3.71KK206 pKa = 11.44VRR208 pKa = 11.84SAVRR212 pKa = 11.84AGQDD216 pKa = 3.1VSSFFDD222 pKa = 4.03TIRR225 pKa = 11.84HH226 pKa = 5.44GGILQLRR233 pKa = 11.84VMGEE237 pKa = 4.37LKK239 pKa = 10.16TSSTGNMQIQILVFVKK255 pKa = 10.26PGPDD259 pKa = 3.4FQFSTLIPPTVKK271 pKa = 10.51VGPTPSQVPAVIASLLSSAVAVRR294 pKa = 11.84RR295 pKa = 11.84CSATLSPVRR304 pKa = 11.84TLLVVSASVTPNKK317 pKa = 9.26MWFYY321 pKa = 11.44GCASSQGDD329 pKa = 3.25WCHH332 pKa = 5.16NTRR335 pKa = 11.84SYY337 pKa = 8.6VTKK340 pKa = 10.64DD341 pKa = 3.27PIITHH346 pKa = 5.63SPEE349 pKa = 3.34ISLLRR354 pKa = 11.84FPTVAKK360 pKa = 10.48NDD362 pKa = 3.49FSGYY366 pKa = 8.7VCDD369 pKa = 4.52VRR371 pKa = 11.84GATGNEE377 pKa = 4.36PIAQIAPKK385 pKa = 10.26GDD387 pKa = 3.32LGKK390 pKa = 10.79VVDD393 pKa = 4.82PFDD396 pKa = 5.14CNVLILEE403 pKa = 4.52NVVEE407 pKa = 4.27ANEE410 pKa = 4.14IRR412 pKa = 11.84GTEE415 pKa = 3.83YY416 pKa = 10.85DD417 pKa = 3.85APDD420 pKa = 3.8SVWIAEE426 pKa = 4.4EE427 pKa = 3.93NLSADD432 pKa = 3.78RR433 pKa = 11.84SEE435 pKa = 4.59VATVVHH441 pKa = 6.18PVGMWEE447 pKa = 5.05LPDD450 pKa = 3.68QEE452 pKa = 5.84LVFADD457 pKa = 3.67SAFMKK462 pKa = 9.09DD463 pKa = 2.64TWHH466 pKa = 6.72RR467 pKa = 11.84TLKK470 pKa = 10.19WPVNSVIDD478 pKa = 4.09RR479 pKa = 11.84EE480 pKa = 4.52SMTAKK485 pKa = 10.53LNLNFMPANAPVCWRR500 pKa = 11.84AIRR503 pKa = 11.84PVALDD508 pKa = 3.46LFIKK512 pKa = 10.56KK513 pKa = 9.65EE514 pKa = 3.9YY515 pKa = 10.87LNLKK519 pKa = 8.95PATKK523 pKa = 9.99EE524 pKa = 4.18SVFLFCPIKK533 pKa = 10.68PGTLDD538 pKa = 3.3AASIEE543 pKa = 4.23TGKK546 pKa = 11.01YY547 pKa = 10.16GLPTYY552 pKa = 9.45QTDD555 pKa = 3.47DD556 pKa = 3.2VSRR559 pKa = 11.84YY560 pKa = 8.85LAYY563 pKa = 10.63GNAQGLLTSEE573 pKa = 4.44DD574 pKa = 3.39ALLFEE579 pKa = 5.79LYY581 pKa = 10.1SVPLGTPVRR590 pKa = 11.84YY591 pKa = 10.0LKK593 pKa = 9.95MYY595 pKa = 9.11SSGMITTNASDD606 pKa = 3.36TTLILDD612 pKa = 4.04KK613 pKa = 11.25SDD615 pKa = 4.5DD616 pKa = 3.6YY617 pKa = 11.89DD618 pKa = 3.46VRR620 pKa = 11.84FVQKK624 pKa = 9.94IAASAAIPALPTHH637 pKa = 5.91STWASQVHH645 pKa = 5.92IANVVGKK652 pKa = 10.18RR653 pKa = 11.84EE654 pKa = 3.78NAMLRR659 pKa = 11.84EE660 pKa = 4.33SVTALDD666 pKa = 4.49RR667 pKa = 11.84KK668 pKa = 10.24IEE670 pKa = 4.08LLTSLLRR677 pKa = 11.84KK678 pKa = 9.53
Molecular weight: 74.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLZ2|A0A1L3KLZ2_9VIRU Uncharacterized protein OS=Hubei myriapoda virus 2 OX=1922931 PE=4 SV=1
MM1 pKa = 7.69AFQYY5 pKa = 11.01SAFLKK10 pKa = 10.89LMTILQNLSDD20 pKa = 4.59DD21 pKa = 3.91GKK23 pKa = 11.02LYY25 pKa = 10.77KK26 pKa = 10.27PSNLRR31 pKa = 11.84LLFYY35 pKa = 9.7TSPPAFWTIEE45 pKa = 3.99WIRR48 pKa = 11.84TFGFPAWLFPVFGSLFPDD66 pKa = 4.2NIHH69 pKa = 6.43ALSSKK74 pKa = 10.37SYY76 pKa = 8.38WVPSFTSLFSQVAVFWSHH94 pKa = 6.3GFTIDD99 pKa = 5.16DD100 pKa = 3.43LHH102 pKa = 7.59KK103 pKa = 11.18YY104 pKa = 10.25SDD106 pKa = 3.65HH107 pKa = 6.23QLVAKK112 pKa = 10.22ILTNFPIVTPKK123 pKa = 10.46HH124 pKa = 6.49DD125 pKa = 4.37FVTTMYY131 pKa = 9.45TCVGFFRR138 pKa = 11.84LQITAHH144 pKa = 6.69EE145 pKa = 4.7DD146 pKa = 2.89ADD148 pKa = 3.55IVRR151 pKa = 11.84RR152 pKa = 11.84HH153 pKa = 6.39LLPLLHH159 pKa = 7.18PGTRR163 pKa = 11.84HH164 pKa = 6.08CGCVQCFPAIKK175 pKa = 9.82KK176 pKa = 10.63YY177 pKa = 10.42YY178 pKa = 9.89DD179 pKa = 3.4LRR181 pKa = 11.84RR182 pKa = 11.84DD183 pKa = 3.17SSNIFLKK190 pKa = 10.86GIPSVYY196 pKa = 9.88LKK198 pKa = 10.59HH199 pKa = 6.01HH200 pKa = 5.67TAHH203 pKa = 6.58PHH205 pKa = 5.04FHH207 pKa = 7.0PFDD210 pKa = 4.3AFTCAVPGALSRR222 pKa = 11.84RR223 pKa = 11.84VVAPPTFRR231 pKa = 11.84KK232 pKa = 9.62ILSLFYY238 pKa = 9.02TKK240 pKa = 10.35CYY242 pKa = 10.41GALQSLGRR250 pKa = 11.84EE251 pKa = 4.04VHH253 pKa = 5.73TSIDD257 pKa = 3.42ALSNPLCDD265 pKa = 3.46GLEE268 pKa = 4.04PQRR271 pKa = 11.84LEE273 pKa = 4.49DD274 pKa = 4.62LSDD277 pKa = 3.09THH279 pKa = 7.65EE280 pKa = 4.3EE281 pKa = 4.45VFCPAMRR288 pKa = 11.84YY289 pKa = 8.84FVVHH293 pKa = 6.6HH294 pKa = 6.77LMTTTEE300 pKa = 4.1RR301 pKa = 11.84QFALNMTWDD310 pKa = 3.6LFLNFFNIPIASVAVEE326 pKa = 3.96LSVSRR331 pKa = 11.84WKK333 pKa = 10.95VFSEE337 pKa = 4.18KK338 pKa = 9.73IVRR341 pKa = 11.84SFWVPEE347 pKa = 3.73EE348 pKa = 3.88EE349 pKa = 4.4LYY351 pKa = 11.19EE352 pKa = 4.31FIIFANHH359 pKa = 6.67LCVCPGDD366 pKa = 4.99FIGEE370 pKa = 4.17PNFVKK375 pKa = 10.89FMDD378 pKa = 3.68AVTRR382 pKa = 11.84VCIQLSVLINLRR394 pKa = 11.84IALCSFVHH402 pKa = 6.9ADD404 pKa = 3.57VLADD408 pKa = 3.9TAALASRR415 pKa = 11.84IEE417 pKa = 4.36DD418 pKa = 3.8NEE420 pKa = 3.84WTPNIFFQLFRR431 pKa = 11.84TKK433 pKa = 9.68QQPRR437 pKa = 11.84SLMQLSHH444 pKa = 6.58MATMGEE450 pKa = 4.51FISTHH455 pKa = 5.99SEE457 pKa = 3.56MSEE460 pKa = 4.03NPDD463 pKa = 3.03MDD465 pKa = 3.84VVGG468 pKa = 4.14
MM1 pKa = 7.69AFQYY5 pKa = 11.01SAFLKK10 pKa = 10.89LMTILQNLSDD20 pKa = 4.59DD21 pKa = 3.91GKK23 pKa = 11.02LYY25 pKa = 10.77KK26 pKa = 10.27PSNLRR31 pKa = 11.84LLFYY35 pKa = 9.7TSPPAFWTIEE45 pKa = 3.99WIRR48 pKa = 11.84TFGFPAWLFPVFGSLFPDD66 pKa = 4.2NIHH69 pKa = 6.43ALSSKK74 pKa = 10.37SYY76 pKa = 8.38WVPSFTSLFSQVAVFWSHH94 pKa = 6.3GFTIDD99 pKa = 5.16DD100 pKa = 3.43LHH102 pKa = 7.59KK103 pKa = 11.18YY104 pKa = 10.25SDD106 pKa = 3.65HH107 pKa = 6.23QLVAKK112 pKa = 10.22ILTNFPIVTPKK123 pKa = 10.46HH124 pKa = 6.49DD125 pKa = 4.37FVTTMYY131 pKa = 9.45TCVGFFRR138 pKa = 11.84LQITAHH144 pKa = 6.69EE145 pKa = 4.7DD146 pKa = 2.89ADD148 pKa = 3.55IVRR151 pKa = 11.84RR152 pKa = 11.84HH153 pKa = 6.39LLPLLHH159 pKa = 7.18PGTRR163 pKa = 11.84HH164 pKa = 6.08CGCVQCFPAIKK175 pKa = 9.82KK176 pKa = 10.63YY177 pKa = 10.42YY178 pKa = 9.89DD179 pKa = 3.4LRR181 pKa = 11.84RR182 pKa = 11.84DD183 pKa = 3.17SSNIFLKK190 pKa = 10.86GIPSVYY196 pKa = 9.88LKK198 pKa = 10.59HH199 pKa = 6.01HH200 pKa = 5.67TAHH203 pKa = 6.58PHH205 pKa = 5.04FHH207 pKa = 7.0PFDD210 pKa = 4.3AFTCAVPGALSRR222 pKa = 11.84RR223 pKa = 11.84VVAPPTFRR231 pKa = 11.84KK232 pKa = 9.62ILSLFYY238 pKa = 9.02TKK240 pKa = 10.35CYY242 pKa = 10.41GALQSLGRR250 pKa = 11.84EE251 pKa = 4.04VHH253 pKa = 5.73TSIDD257 pKa = 3.42ALSNPLCDD265 pKa = 3.46GLEE268 pKa = 4.04PQRR271 pKa = 11.84LEE273 pKa = 4.49DD274 pKa = 4.62LSDD277 pKa = 3.09THH279 pKa = 7.65EE280 pKa = 4.3EE281 pKa = 4.45VFCPAMRR288 pKa = 11.84YY289 pKa = 8.84FVVHH293 pKa = 6.6HH294 pKa = 6.77LMTTTEE300 pKa = 4.1RR301 pKa = 11.84QFALNMTWDD310 pKa = 3.6LFLNFFNIPIASVAVEE326 pKa = 3.96LSVSRR331 pKa = 11.84WKK333 pKa = 10.95VFSEE337 pKa = 4.18KK338 pKa = 9.73IVRR341 pKa = 11.84SFWVPEE347 pKa = 3.73EE348 pKa = 3.88EE349 pKa = 4.4LYY351 pKa = 11.19EE352 pKa = 4.31FIIFANHH359 pKa = 6.67LCVCPGDD366 pKa = 4.99FIGEE370 pKa = 4.17PNFVKK375 pKa = 10.89FMDD378 pKa = 3.68AVTRR382 pKa = 11.84VCIQLSVLINLRR394 pKa = 11.84IALCSFVHH402 pKa = 6.9ADD404 pKa = 3.57VLADD408 pKa = 3.9TAALASRR415 pKa = 11.84IEE417 pKa = 4.36DD418 pKa = 3.8NEE420 pKa = 3.84WTPNIFFQLFRR431 pKa = 11.84TKK433 pKa = 9.68QQPRR437 pKa = 11.84SLMQLSHH444 pKa = 6.58MATMGEE450 pKa = 4.51FISTHH455 pKa = 5.99SEE457 pKa = 3.56MSEE460 pKa = 4.03NPDD463 pKa = 3.03MDD465 pKa = 3.84VVGG468 pKa = 4.14
Molecular weight: 53.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3244 |
468 |
2098 |
1081.3 |
121.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.905 ± 0.655 | 1.819 ± 0.204 |
6.196 ± 0.393 | 5.21 ± 0.461 |
5.025 ± 1.002 | 5.333 ± 0.475 |
2.682 ± 0.501 | 5.949 ± 0.199 |
4.84 ± 0.503 | 9.895 ± 0.247 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.497 ± 0.149 | 4.192 ± 0.283 |
5.425 ± 0.681 | 3.422 ± 0.297 |
5.456 ± 0.502 | 7.398 ± 0.429 |
5.672 ± 0.64 | 7.429 ± 0.373 |
1.541 ± 0.16 | 3.113 ± 0.083 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
