
Magnaporthe oryzae ourmia-like virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Miaviricetes; Ourlivirales; Botourmiaviridae; Magoulivirus; Magnaporthe magoulivirus 1
Average proteome isoelectric point is 9.28
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M4HCZ9|A0A1M4HCZ9_9VIRU RNA dependent RNA polymerase OS=Magnaporthe oryzae ourmia-like virus OX=1858671 GN=RdRp PE=4 SV=1
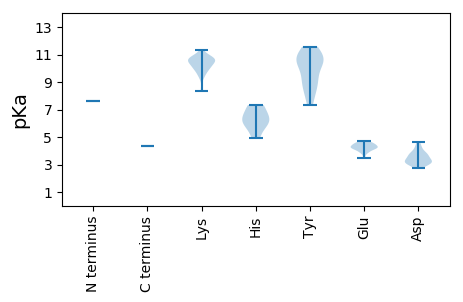
MM1 pKa = 7.63SYY3 pKa = 10.66SEE5 pKa = 4.55SDD7 pKa = 3.07GSAACNSAGRR17 pKa = 11.84VAVPSLQMRR26 pKa = 11.84NLVSFSGLLSEE37 pKa = 4.7TFRR40 pKa = 11.84VDD42 pKa = 3.3LRR44 pKa = 11.84VPEE47 pKa = 4.13TLGDD51 pKa = 3.63STRR54 pKa = 11.84EE55 pKa = 3.83LKK57 pKa = 10.42EE58 pKa = 3.48FCGRR62 pKa = 11.84LLEE65 pKa = 4.54PSGSHH70 pKa = 5.82PWAHH74 pKa = 7.2ILRR77 pKa = 11.84PLPAVDD83 pKa = 3.85RR84 pKa = 11.84LSIAGSLFSFSKK96 pKa = 9.66TLPSSDD102 pKa = 4.62PDD104 pKa = 3.19CDD106 pKa = 3.25AYY108 pKa = 10.58VRR110 pKa = 11.84HH111 pKa = 5.9MSVPGVKK118 pKa = 9.45TPDD121 pKa = 3.0GYY123 pKa = 11.57LRR125 pKa = 11.84FVRR128 pKa = 11.84KK129 pKa = 10.05LIMRR133 pKa = 11.84EE134 pKa = 4.23FPVGWDD140 pKa = 3.12KK141 pKa = 11.32GWRR144 pKa = 11.84STYY147 pKa = 10.79LGTTPTSSSALGSSKK162 pKa = 10.9GKK164 pKa = 10.65GGARR168 pKa = 11.84YY169 pKa = 8.06QLRR172 pKa = 11.84GSRR175 pKa = 11.84QWFLKK180 pKa = 10.21SCMGLEE186 pKa = 4.24RR187 pKa = 11.84EE188 pKa = 4.46LLLDD192 pKa = 3.57PVRR195 pKa = 11.84KK196 pKa = 10.09VGIARR201 pKa = 11.84CDD203 pKa = 3.36GKK205 pKa = 10.63SRR207 pKa = 11.84RR208 pKa = 11.84VTQSPAEE215 pKa = 4.21SVVLSPLHH223 pKa = 4.95TLIYY227 pKa = 10.4NHH229 pKa = 7.33LSRR232 pKa = 11.84KK233 pKa = 9.34DD234 pKa = 3.04WLLRR238 pKa = 11.84GEE240 pKa = 4.08ATPDD244 pKa = 3.05KK245 pKa = 10.76FADD248 pKa = 4.28FCQKK252 pKa = 10.56DD253 pKa = 3.67GEE255 pKa = 4.69VFVSGDD261 pKa = 3.54YY262 pKa = 10.98EE263 pKa = 4.32SASDD267 pKa = 3.79NLSIEE272 pKa = 4.03AAEE275 pKa = 4.4TVLSAIFAKK284 pKa = 10.38ARR286 pKa = 11.84YY287 pKa = 8.32IPQAIRR293 pKa = 11.84RR294 pKa = 11.84VAMDD298 pKa = 3.31SLRR301 pKa = 11.84CVLVSEE307 pKa = 4.58GAVGIQARR315 pKa = 11.84GQLMGNFLCFPLLCLQNYY333 pKa = 9.16AAFRR337 pKa = 11.84YY338 pKa = 9.25LAGNYY343 pKa = 7.31PVRR346 pKa = 11.84INGDD350 pKa = 3.8DD351 pKa = 3.22IVFRR355 pKa = 11.84APEE358 pKa = 3.86HH359 pKa = 6.22VRR361 pKa = 11.84ARR363 pKa = 11.84WAAGVQALGLTLSVGKK379 pKa = 8.33TFVHH383 pKa = 6.56KK384 pKa = 10.64RR385 pKa = 11.84FFSLNSTYY393 pKa = 10.23FRR395 pKa = 11.84ARR397 pKa = 11.84TKK399 pKa = 10.87NGVKK403 pKa = 10.02GVPIVRR409 pKa = 11.84ATLFYY414 pKa = 10.77KK415 pKa = 10.67GVEE418 pKa = 4.3SADD421 pKa = 3.86SIVGRR426 pKa = 11.84FSSVGSFFPVATSSMLRR443 pKa = 11.84VRR445 pKa = 11.84ILQAMQKK452 pKa = 9.96EE453 pKa = 4.48IRR455 pKa = 11.84CTQRR459 pKa = 11.84SVRR462 pKa = 11.84RR463 pKa = 11.84GLGLNATPAEE473 pKa = 4.18LARR476 pKa = 11.84SGLLVRR482 pKa = 11.84EE483 pKa = 4.28KK484 pKa = 10.51FYY486 pKa = 11.54SNLPRR491 pKa = 11.84EE492 pKa = 4.27SAVPSKK498 pKa = 10.7LAVSGALPPGWVKK511 pKa = 10.04VRR513 pKa = 11.84PLWTGMTLVEE523 pKa = 4.47DD524 pKa = 4.22PEE526 pKa = 4.48FKK528 pKa = 10.62KK529 pKa = 10.83EE530 pKa = 4.25LVQHH534 pKa = 6.45CWNSVVSRR542 pKa = 11.84VEE544 pKa = 3.95TQDD547 pKa = 2.76YY548 pKa = 8.89WNVVRR553 pKa = 11.84SGGCPYY559 pKa = 10.76RR560 pKa = 11.84STSGQAYY567 pKa = 9.07RR568 pKa = 11.84RR569 pKa = 11.84KK570 pKa = 10.4AKK572 pKa = 10.07LLKK575 pKa = 10.54LSVNATRR582 pKa = 11.84RR583 pKa = 11.84YY584 pKa = 10.23LEE586 pKa = 4.16GRR588 pKa = 11.84LGAVPPRR595 pKa = 11.84KK596 pKa = 9.7KK597 pKa = 10.73GVWQRR602 pKa = 11.84ADD604 pKa = 3.14RR605 pKa = 4.36
MM1 pKa = 7.63SYY3 pKa = 10.66SEE5 pKa = 4.55SDD7 pKa = 3.07GSAACNSAGRR17 pKa = 11.84VAVPSLQMRR26 pKa = 11.84NLVSFSGLLSEE37 pKa = 4.7TFRR40 pKa = 11.84VDD42 pKa = 3.3LRR44 pKa = 11.84VPEE47 pKa = 4.13TLGDD51 pKa = 3.63STRR54 pKa = 11.84EE55 pKa = 3.83LKK57 pKa = 10.42EE58 pKa = 3.48FCGRR62 pKa = 11.84LLEE65 pKa = 4.54PSGSHH70 pKa = 5.82PWAHH74 pKa = 7.2ILRR77 pKa = 11.84PLPAVDD83 pKa = 3.85RR84 pKa = 11.84LSIAGSLFSFSKK96 pKa = 9.66TLPSSDD102 pKa = 4.62PDD104 pKa = 3.19CDD106 pKa = 3.25AYY108 pKa = 10.58VRR110 pKa = 11.84HH111 pKa = 5.9MSVPGVKK118 pKa = 9.45TPDD121 pKa = 3.0GYY123 pKa = 11.57LRR125 pKa = 11.84FVRR128 pKa = 11.84KK129 pKa = 10.05LIMRR133 pKa = 11.84EE134 pKa = 4.23FPVGWDD140 pKa = 3.12KK141 pKa = 11.32GWRR144 pKa = 11.84STYY147 pKa = 10.79LGTTPTSSSALGSSKK162 pKa = 10.9GKK164 pKa = 10.65GGARR168 pKa = 11.84YY169 pKa = 8.06QLRR172 pKa = 11.84GSRR175 pKa = 11.84QWFLKK180 pKa = 10.21SCMGLEE186 pKa = 4.24RR187 pKa = 11.84EE188 pKa = 4.46LLLDD192 pKa = 3.57PVRR195 pKa = 11.84KK196 pKa = 10.09VGIARR201 pKa = 11.84CDD203 pKa = 3.36GKK205 pKa = 10.63SRR207 pKa = 11.84RR208 pKa = 11.84VTQSPAEE215 pKa = 4.21SVVLSPLHH223 pKa = 4.95TLIYY227 pKa = 10.4NHH229 pKa = 7.33LSRR232 pKa = 11.84KK233 pKa = 9.34DD234 pKa = 3.04WLLRR238 pKa = 11.84GEE240 pKa = 4.08ATPDD244 pKa = 3.05KK245 pKa = 10.76FADD248 pKa = 4.28FCQKK252 pKa = 10.56DD253 pKa = 3.67GEE255 pKa = 4.69VFVSGDD261 pKa = 3.54YY262 pKa = 10.98EE263 pKa = 4.32SASDD267 pKa = 3.79NLSIEE272 pKa = 4.03AAEE275 pKa = 4.4TVLSAIFAKK284 pKa = 10.38ARR286 pKa = 11.84YY287 pKa = 8.32IPQAIRR293 pKa = 11.84RR294 pKa = 11.84VAMDD298 pKa = 3.31SLRR301 pKa = 11.84CVLVSEE307 pKa = 4.58GAVGIQARR315 pKa = 11.84GQLMGNFLCFPLLCLQNYY333 pKa = 9.16AAFRR337 pKa = 11.84YY338 pKa = 9.25LAGNYY343 pKa = 7.31PVRR346 pKa = 11.84INGDD350 pKa = 3.8DD351 pKa = 3.22IVFRR355 pKa = 11.84APEE358 pKa = 3.86HH359 pKa = 6.22VRR361 pKa = 11.84ARR363 pKa = 11.84WAAGVQALGLTLSVGKK379 pKa = 8.33TFVHH383 pKa = 6.56KK384 pKa = 10.64RR385 pKa = 11.84FFSLNSTYY393 pKa = 10.23FRR395 pKa = 11.84ARR397 pKa = 11.84TKK399 pKa = 10.87NGVKK403 pKa = 10.02GVPIVRR409 pKa = 11.84ATLFYY414 pKa = 10.77KK415 pKa = 10.67GVEE418 pKa = 4.3SADD421 pKa = 3.86SIVGRR426 pKa = 11.84FSSVGSFFPVATSSMLRR443 pKa = 11.84VRR445 pKa = 11.84ILQAMQKK452 pKa = 9.96EE453 pKa = 4.48IRR455 pKa = 11.84CTQRR459 pKa = 11.84SVRR462 pKa = 11.84RR463 pKa = 11.84GLGLNATPAEE473 pKa = 4.18LARR476 pKa = 11.84SGLLVRR482 pKa = 11.84EE483 pKa = 4.28KK484 pKa = 10.51FYY486 pKa = 11.54SNLPRR491 pKa = 11.84EE492 pKa = 4.27SAVPSKK498 pKa = 10.7LAVSGALPPGWVKK511 pKa = 10.04VRR513 pKa = 11.84PLWTGMTLVEE523 pKa = 4.47DD524 pKa = 4.22PEE526 pKa = 4.48FKK528 pKa = 10.62KK529 pKa = 10.83EE530 pKa = 4.25LVQHH534 pKa = 6.45CWNSVVSRR542 pKa = 11.84VEE544 pKa = 3.95TQDD547 pKa = 2.76YY548 pKa = 8.89WNVVRR553 pKa = 11.84SGGCPYY559 pKa = 10.76RR560 pKa = 11.84STSGQAYY567 pKa = 9.07RR568 pKa = 11.84RR569 pKa = 11.84KK570 pKa = 10.4AKK572 pKa = 10.07LLKK575 pKa = 10.54LSVNATRR582 pKa = 11.84RR583 pKa = 11.84YY584 pKa = 10.23LEE586 pKa = 4.16GRR588 pKa = 11.84LGAVPPRR595 pKa = 11.84KK596 pKa = 9.7KK597 pKa = 10.73GVWQRR602 pKa = 11.84ADD604 pKa = 3.14RR605 pKa = 4.36
Molecular weight: 67.12 kDa
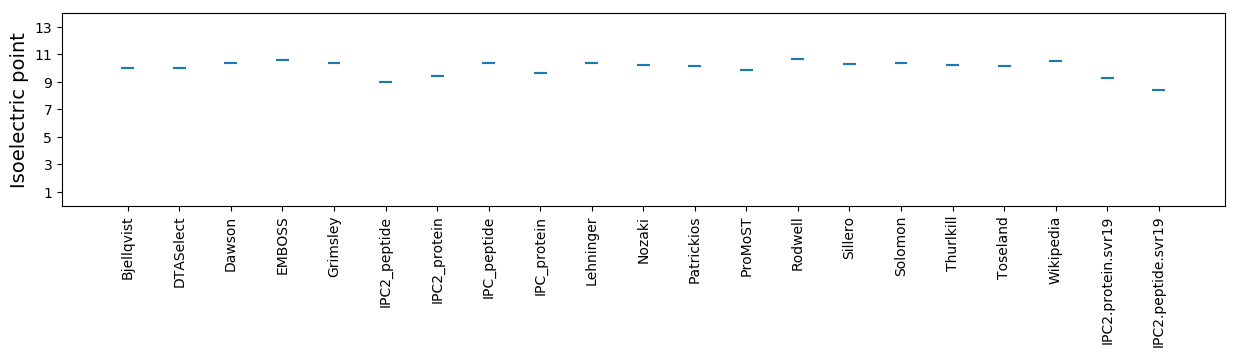
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M4HCZ9|A0A1M4HCZ9_9VIRU RNA dependent RNA polymerase OS=Magnaporthe oryzae ourmia-like virus OX=1858671 GN=RdRp PE=4 SV=1
MM1 pKa = 7.63SYY3 pKa = 10.66SEE5 pKa = 4.55SDD7 pKa = 3.07GSAACNSAGRR17 pKa = 11.84VAVPSLQMRR26 pKa = 11.84NLVSFSGLLSEE37 pKa = 4.7TFRR40 pKa = 11.84VDD42 pKa = 3.3LRR44 pKa = 11.84VPEE47 pKa = 4.13TLGDD51 pKa = 3.63STRR54 pKa = 11.84EE55 pKa = 3.83LKK57 pKa = 10.42EE58 pKa = 3.48FCGRR62 pKa = 11.84LLEE65 pKa = 4.54PSGSHH70 pKa = 5.82PWAHH74 pKa = 7.2ILRR77 pKa = 11.84PLPAVDD83 pKa = 3.85RR84 pKa = 11.84LSIAGSLFSFSKK96 pKa = 9.66TLPSSDD102 pKa = 4.62PDD104 pKa = 3.19CDD106 pKa = 3.25AYY108 pKa = 10.58VRR110 pKa = 11.84HH111 pKa = 5.9MSVPGVKK118 pKa = 9.45TPDD121 pKa = 3.0GYY123 pKa = 11.57LRR125 pKa = 11.84FVRR128 pKa = 11.84KK129 pKa = 10.05LIMRR133 pKa = 11.84EE134 pKa = 4.23FPVGWDD140 pKa = 3.12KK141 pKa = 11.32GWRR144 pKa = 11.84STYY147 pKa = 10.79LGTTPTSSSALGSSKK162 pKa = 10.9GKK164 pKa = 10.65GGARR168 pKa = 11.84YY169 pKa = 8.06QLRR172 pKa = 11.84GSRR175 pKa = 11.84QWFLKK180 pKa = 10.21SCMGLEE186 pKa = 4.24RR187 pKa = 11.84EE188 pKa = 4.46LLLDD192 pKa = 3.57PVRR195 pKa = 11.84KK196 pKa = 10.09VGIARR201 pKa = 11.84CDD203 pKa = 3.36GKK205 pKa = 10.63SRR207 pKa = 11.84RR208 pKa = 11.84VTQSPAEE215 pKa = 4.21SVVLSPLHH223 pKa = 4.95TLIYY227 pKa = 10.4NHH229 pKa = 7.33LSRR232 pKa = 11.84KK233 pKa = 9.34DD234 pKa = 3.04WLLRR238 pKa = 11.84GEE240 pKa = 4.08ATPDD244 pKa = 3.05KK245 pKa = 10.76FADD248 pKa = 4.28FCQKK252 pKa = 10.56DD253 pKa = 3.67GEE255 pKa = 4.69VFVSGDD261 pKa = 3.54YY262 pKa = 10.98EE263 pKa = 4.32SASDD267 pKa = 3.79NLSIEE272 pKa = 4.03AAEE275 pKa = 4.4TVLSAIFAKK284 pKa = 10.38ARR286 pKa = 11.84YY287 pKa = 8.32IPQAIRR293 pKa = 11.84RR294 pKa = 11.84VAMDD298 pKa = 3.31SLRR301 pKa = 11.84CVLVSEE307 pKa = 4.58GAVGIQARR315 pKa = 11.84GQLMGNFLCFPLLCLQNYY333 pKa = 9.16AAFRR337 pKa = 11.84YY338 pKa = 9.25LAGNYY343 pKa = 7.31PVRR346 pKa = 11.84INGDD350 pKa = 3.8DD351 pKa = 3.22IVFRR355 pKa = 11.84APEE358 pKa = 3.86HH359 pKa = 6.22VRR361 pKa = 11.84ARR363 pKa = 11.84WAAGVQALGLTLSVGKK379 pKa = 8.33TFVHH383 pKa = 6.56KK384 pKa = 10.64RR385 pKa = 11.84FFSLNSTYY393 pKa = 10.23FRR395 pKa = 11.84ARR397 pKa = 11.84TKK399 pKa = 10.87NGVKK403 pKa = 10.02GVPIVRR409 pKa = 11.84ATLFYY414 pKa = 10.77KK415 pKa = 10.67GVEE418 pKa = 4.3SADD421 pKa = 3.86SIVGRR426 pKa = 11.84FSSVGSFFPVATSSMLRR443 pKa = 11.84VRR445 pKa = 11.84ILQAMQKK452 pKa = 9.96EE453 pKa = 4.48IRR455 pKa = 11.84CTQRR459 pKa = 11.84SVRR462 pKa = 11.84RR463 pKa = 11.84GLGLNATPAEE473 pKa = 4.18LARR476 pKa = 11.84SGLLVRR482 pKa = 11.84EE483 pKa = 4.28KK484 pKa = 10.51FYY486 pKa = 11.54SNLPRR491 pKa = 11.84EE492 pKa = 4.27SAVPSKK498 pKa = 10.7LAVSGALPPGWVKK511 pKa = 10.04VRR513 pKa = 11.84PLWTGMTLVEE523 pKa = 4.47DD524 pKa = 4.22PEE526 pKa = 4.48FKK528 pKa = 10.62KK529 pKa = 10.83EE530 pKa = 4.25LVQHH534 pKa = 6.45CWNSVVSRR542 pKa = 11.84VEE544 pKa = 3.95TQDD547 pKa = 2.76YY548 pKa = 8.89WNVVRR553 pKa = 11.84SGGCPYY559 pKa = 10.76RR560 pKa = 11.84STSGQAYY567 pKa = 9.07RR568 pKa = 11.84RR569 pKa = 11.84KK570 pKa = 10.4AKK572 pKa = 10.07LLKK575 pKa = 10.54LSVNATRR582 pKa = 11.84RR583 pKa = 11.84YY584 pKa = 10.23LEE586 pKa = 4.16GRR588 pKa = 11.84LGAVPPRR595 pKa = 11.84KK596 pKa = 9.7KK597 pKa = 10.73GVWQRR602 pKa = 11.84ADD604 pKa = 3.14RR605 pKa = 4.36
MM1 pKa = 7.63SYY3 pKa = 10.66SEE5 pKa = 4.55SDD7 pKa = 3.07GSAACNSAGRR17 pKa = 11.84VAVPSLQMRR26 pKa = 11.84NLVSFSGLLSEE37 pKa = 4.7TFRR40 pKa = 11.84VDD42 pKa = 3.3LRR44 pKa = 11.84VPEE47 pKa = 4.13TLGDD51 pKa = 3.63STRR54 pKa = 11.84EE55 pKa = 3.83LKK57 pKa = 10.42EE58 pKa = 3.48FCGRR62 pKa = 11.84LLEE65 pKa = 4.54PSGSHH70 pKa = 5.82PWAHH74 pKa = 7.2ILRR77 pKa = 11.84PLPAVDD83 pKa = 3.85RR84 pKa = 11.84LSIAGSLFSFSKK96 pKa = 9.66TLPSSDD102 pKa = 4.62PDD104 pKa = 3.19CDD106 pKa = 3.25AYY108 pKa = 10.58VRR110 pKa = 11.84HH111 pKa = 5.9MSVPGVKK118 pKa = 9.45TPDD121 pKa = 3.0GYY123 pKa = 11.57LRR125 pKa = 11.84FVRR128 pKa = 11.84KK129 pKa = 10.05LIMRR133 pKa = 11.84EE134 pKa = 4.23FPVGWDD140 pKa = 3.12KK141 pKa = 11.32GWRR144 pKa = 11.84STYY147 pKa = 10.79LGTTPTSSSALGSSKK162 pKa = 10.9GKK164 pKa = 10.65GGARR168 pKa = 11.84YY169 pKa = 8.06QLRR172 pKa = 11.84GSRR175 pKa = 11.84QWFLKK180 pKa = 10.21SCMGLEE186 pKa = 4.24RR187 pKa = 11.84EE188 pKa = 4.46LLLDD192 pKa = 3.57PVRR195 pKa = 11.84KK196 pKa = 10.09VGIARR201 pKa = 11.84CDD203 pKa = 3.36GKK205 pKa = 10.63SRR207 pKa = 11.84RR208 pKa = 11.84VTQSPAEE215 pKa = 4.21SVVLSPLHH223 pKa = 4.95TLIYY227 pKa = 10.4NHH229 pKa = 7.33LSRR232 pKa = 11.84KK233 pKa = 9.34DD234 pKa = 3.04WLLRR238 pKa = 11.84GEE240 pKa = 4.08ATPDD244 pKa = 3.05KK245 pKa = 10.76FADD248 pKa = 4.28FCQKK252 pKa = 10.56DD253 pKa = 3.67GEE255 pKa = 4.69VFVSGDD261 pKa = 3.54YY262 pKa = 10.98EE263 pKa = 4.32SASDD267 pKa = 3.79NLSIEE272 pKa = 4.03AAEE275 pKa = 4.4TVLSAIFAKK284 pKa = 10.38ARR286 pKa = 11.84YY287 pKa = 8.32IPQAIRR293 pKa = 11.84RR294 pKa = 11.84VAMDD298 pKa = 3.31SLRR301 pKa = 11.84CVLVSEE307 pKa = 4.58GAVGIQARR315 pKa = 11.84GQLMGNFLCFPLLCLQNYY333 pKa = 9.16AAFRR337 pKa = 11.84YY338 pKa = 9.25LAGNYY343 pKa = 7.31PVRR346 pKa = 11.84INGDD350 pKa = 3.8DD351 pKa = 3.22IVFRR355 pKa = 11.84APEE358 pKa = 3.86HH359 pKa = 6.22VRR361 pKa = 11.84ARR363 pKa = 11.84WAAGVQALGLTLSVGKK379 pKa = 8.33TFVHH383 pKa = 6.56KK384 pKa = 10.64RR385 pKa = 11.84FFSLNSTYY393 pKa = 10.23FRR395 pKa = 11.84ARR397 pKa = 11.84TKK399 pKa = 10.87NGVKK403 pKa = 10.02GVPIVRR409 pKa = 11.84ATLFYY414 pKa = 10.77KK415 pKa = 10.67GVEE418 pKa = 4.3SADD421 pKa = 3.86SIVGRR426 pKa = 11.84FSSVGSFFPVATSSMLRR443 pKa = 11.84VRR445 pKa = 11.84ILQAMQKK452 pKa = 9.96EE453 pKa = 4.48IRR455 pKa = 11.84CTQRR459 pKa = 11.84SVRR462 pKa = 11.84RR463 pKa = 11.84GLGLNATPAEE473 pKa = 4.18LARR476 pKa = 11.84SGLLVRR482 pKa = 11.84EE483 pKa = 4.28KK484 pKa = 10.51FYY486 pKa = 11.54SNLPRR491 pKa = 11.84EE492 pKa = 4.27SAVPSKK498 pKa = 10.7LAVSGALPPGWVKK511 pKa = 10.04VRR513 pKa = 11.84PLWTGMTLVEE523 pKa = 4.47DD524 pKa = 4.22PEE526 pKa = 4.48FKK528 pKa = 10.62KK529 pKa = 10.83EE530 pKa = 4.25LVQHH534 pKa = 6.45CWNSVVSRR542 pKa = 11.84VEE544 pKa = 3.95TQDD547 pKa = 2.76YY548 pKa = 8.89WNVVRR553 pKa = 11.84SGGCPYY559 pKa = 10.76RR560 pKa = 11.84STSGQAYY567 pKa = 9.07RR568 pKa = 11.84RR569 pKa = 11.84KK570 pKa = 10.4AKK572 pKa = 10.07LLKK575 pKa = 10.54LSVNATRR582 pKa = 11.84RR583 pKa = 11.84YY584 pKa = 10.23LEE586 pKa = 4.16GRR588 pKa = 11.84LGAVPPRR595 pKa = 11.84KK596 pKa = 9.7KK597 pKa = 10.73GVWQRR602 pKa = 11.84ADD604 pKa = 3.14RR605 pKa = 4.36
Molecular weight: 67.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
605 |
605 |
605 |
605.0 |
67.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.934 ± 0.0 | 1.983 ± 0.0 |
3.967 ± 0.0 | 4.463 ± 0.0 |
4.298 ± 0.0 | 8.099 ± 0.0 |
1.322 ± 0.0 | 2.645 ± 0.0 |
4.959 ± 0.0 | 10.413 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.653 ± 0.0 | 2.479 ± 0.0 |
5.289 ± 0.0 | 2.81 ± 0.0 |
9.752 ± 0.0 | 10.083 ± 0.0 |
4.298 ± 0.0 | 8.76 ± 0.0 |
1.818 ± 0.0 | 2.975 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
