
Soybean vein necrosis virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Tospoviridae; Orthotospovirus; Soybean vein necrosis orthotospovirus
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
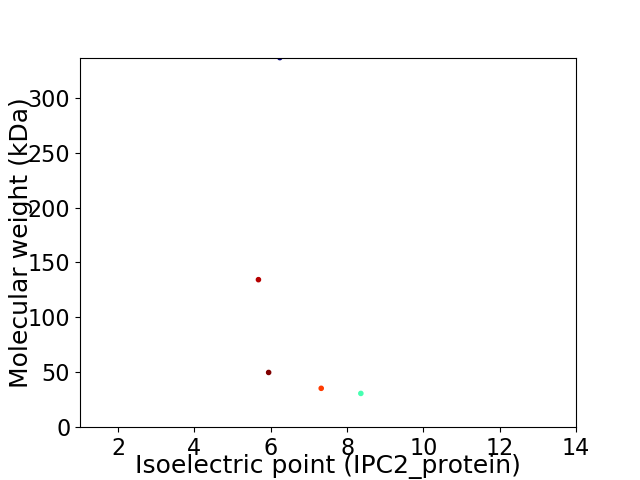
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2Y3N6|F2Y3N6_9VIRU Non-structural protein OS=Soybean vein necrosis virus OX=980895 PE=4 SV=1
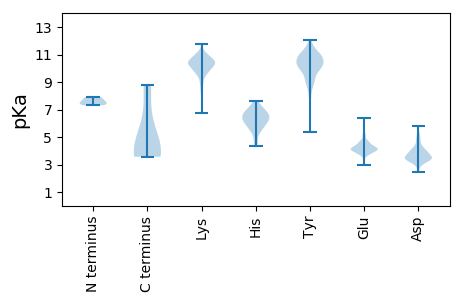
MM1 pKa = 7.91RR2 pKa = 11.84IKK4 pKa = 10.86DD5 pKa = 3.61FFVIMLLFGLNLVFLSIFINASIRR29 pKa = 11.84GDD31 pKa = 3.46HH32 pKa = 6.39EE33 pKa = 4.65VSQEE37 pKa = 3.89TSFEE41 pKa = 4.11EE42 pKa = 4.07LSKK45 pKa = 11.17EE46 pKa = 3.91VMEE49 pKa = 4.29AHH51 pKa = 6.03EE52 pKa = 4.09QGKK55 pKa = 9.67IGKK58 pKa = 8.54SLVSSRR64 pKa = 11.84VEE66 pKa = 3.64AFEE69 pKa = 4.45SFNKK73 pKa = 10.15EE74 pKa = 3.73PIEE77 pKa = 3.94TSRR80 pKa = 11.84RR81 pKa = 11.84PEE83 pKa = 3.67MNDD86 pKa = 4.02RR87 pKa = 11.84ITQTKK92 pKa = 9.63PGIDD96 pKa = 3.43DD97 pKa = 3.44QMIPKK102 pKa = 9.76HH103 pKa = 6.25GEE105 pKa = 4.16GKK107 pKa = 10.16RR108 pKa = 11.84EE109 pKa = 3.94APPPSKK115 pKa = 11.0VDD117 pKa = 3.74LSCANINPEE126 pKa = 3.68NCKK129 pKa = 10.34VVGKK133 pKa = 8.49TPFNFYY139 pKa = 10.59YY140 pKa = 10.31QISDD144 pKa = 3.68SEE146 pKa = 4.33EE147 pKa = 4.25TISCLSSKK155 pKa = 10.82VEE157 pKa = 4.57DD158 pKa = 4.35LKK160 pKa = 11.51ACNPQSMHH168 pKa = 5.85VFTSAPVLPITTILNKK184 pKa = 10.04RR185 pKa = 11.84VLSVGSKK192 pKa = 10.64YY193 pKa = 10.83YY194 pKa = 9.78IQNSIDD200 pKa = 3.44SSSFPVQDD208 pKa = 3.37SGNVQLGLVNIYY220 pKa = 8.46SVRR223 pKa = 11.84LTGEE227 pKa = 3.98CNITKK232 pKa = 10.03VSLTNPFFVTLTSSNKK248 pKa = 6.99EE249 pKa = 3.8TFYY252 pKa = 10.94SIKK255 pKa = 10.48KK256 pKa = 9.94IGADD260 pKa = 3.53DD261 pKa = 3.72VNLIKK266 pKa = 10.76FSGEE270 pKa = 3.81KK271 pKa = 9.39TVEE274 pKa = 4.07LPSDD278 pKa = 3.87SVDD281 pKa = 3.43GNHH284 pKa = 6.49VLLCGDD290 pKa = 4.17KK291 pKa = 10.82VSEE294 pKa = 4.15IPKK297 pKa = 9.43IDD299 pKa = 3.56VNKK302 pKa = 10.28RR303 pKa = 11.84NCLVKK308 pKa = 10.93YY309 pKa = 9.93KK310 pKa = 10.54GKK312 pKa = 10.46RR313 pKa = 11.84YY314 pKa = 8.06QQSACVHH321 pKa = 5.66FNILRR326 pKa = 11.84YY327 pKa = 9.69LLVSIILFVPVSWFLNKK344 pKa = 9.97TKK346 pKa = 11.0DD347 pKa = 5.21SMFLWYY353 pKa = 10.0DD354 pKa = 3.01FLGIIFYY361 pKa = 9.9PILYY365 pKa = 9.1ILNWLWRR372 pKa = 11.84YY373 pKa = 9.47FPMKK377 pKa = 10.32CSNCGNFSLITHH389 pKa = 6.32SCYY392 pKa = 9.73DD393 pKa = 3.3TCVCNKK399 pKa = 9.9SKK401 pKa = 10.93AKK403 pKa = 10.35SEE405 pKa = 4.03HH406 pKa = 6.74AEE408 pKa = 3.75NCPIITGKK416 pKa = 10.48AFSKK420 pKa = 11.07SEE422 pKa = 3.97DD423 pKa = 3.45QDD425 pKa = 3.7VDD427 pKa = 3.82DD428 pKa = 5.73LSVEE432 pKa = 4.24EE433 pKa = 4.69TEE435 pKa = 5.08SLTGSKK441 pKa = 10.06KK442 pKa = 9.79PKK444 pKa = 9.23KK445 pKa = 9.86ASSKK449 pKa = 10.05FRR451 pKa = 11.84TFQLVVNTKK460 pKa = 9.66ISTGFLMFVTKK471 pKa = 10.65VLLGFLIFSQMPKK484 pKa = 9.75TMASKK489 pKa = 8.98TATQTISQTEE499 pKa = 4.24TQVLCVSYY507 pKa = 10.81CSAKK511 pKa = 10.06PGCNKK516 pKa = 9.72FLFKK520 pKa = 11.1NEE522 pKa = 4.06EE523 pKa = 3.96VCIDD527 pKa = 3.85NPKK530 pKa = 10.34LKK532 pKa = 10.31CNCLIRR538 pKa = 11.84DD539 pKa = 3.55GMINEE544 pKa = 4.01EE545 pKa = 4.22AYY547 pKa = 10.03INGTRR552 pKa = 11.84YY553 pKa = 9.72SVSMVDD559 pKa = 2.89NCLDD563 pKa = 3.54RR564 pKa = 11.84RR565 pKa = 11.84CEE567 pKa = 3.76FSSNKK572 pKa = 9.48VEE574 pKa = 5.25DD575 pKa = 4.26LLACRR580 pKa = 11.84LGCGQLKK587 pKa = 10.08KK588 pKa = 10.74LKK590 pKa = 10.26SSKK593 pKa = 10.26LQSKK597 pKa = 9.54ALKK600 pKa = 10.08QGIVPRR606 pKa = 11.84GASISQIYY614 pKa = 10.31SSLRR618 pKa = 11.84LQDD621 pKa = 4.11GYY623 pKa = 10.78IDD625 pKa = 4.09SEE627 pKa = 4.45EE628 pKa = 4.29SLHH631 pKa = 7.34SLGDD635 pKa = 3.6LPEE638 pKa = 4.31NKK640 pKa = 9.71VKK642 pKa = 10.56IEE644 pKa = 4.67DD645 pKa = 3.48EE646 pKa = 4.84DD647 pKa = 3.86IPEE650 pKa = 4.44GVLPRR655 pKa = 11.84QSFVYY660 pKa = 10.59NSIVDD665 pKa = 3.57GKK667 pKa = 9.86YY668 pKa = 9.85RR669 pKa = 11.84YY670 pKa = 9.25LISMDD675 pKa = 4.97AIQSTGYY682 pKa = 9.78VYY684 pKa = 11.43SMNDD688 pKa = 2.79AKK690 pKa = 10.31MAPPQEE696 pKa = 3.72LVVFVKK702 pKa = 10.49EE703 pKa = 4.04AGVTYY708 pKa = 9.9TMKK711 pKa = 10.01TLYY714 pKa = 8.81YY715 pKa = 8.02TAPISATHH723 pKa = 4.55THH725 pKa = 6.12VYY727 pKa = 7.95STCTGNCEE735 pKa = 4.35TCRR738 pKa = 11.84KK739 pKa = 7.05EE740 pKa = 4.37HH741 pKa = 7.07PISGYY746 pKa = 8.75QDD748 pKa = 3.24YY749 pKa = 11.11CIAPTSNWGCEE760 pKa = 4.03EE761 pKa = 4.63LGCLAIGEE769 pKa = 4.64GATCGYY775 pKa = 9.0CRR777 pKa = 11.84NVYY780 pKa = 10.1DD781 pKa = 4.27LSKK784 pKa = 10.35KK785 pKa = 9.69YY786 pKa = 10.14SVRR789 pKa = 11.84QILTSHH795 pKa = 5.46VTVKK799 pKa = 10.58VCFKK803 pKa = 10.59GFAGTGCQTITDD815 pKa = 4.06SVPFQNSYY823 pKa = 10.06YY824 pKa = 10.03QLSIDD829 pKa = 4.63ADD831 pKa = 3.84LHH833 pKa = 7.16NDD835 pKa = 3.45DD836 pKa = 4.57LTAGSRR842 pKa = 11.84VAVDD846 pKa = 3.5PNGMLFKK853 pKa = 11.33GNIANLKK860 pKa = 10.15DD861 pKa = 3.19SSLAFGHH868 pKa = 6.23PQLNEE873 pKa = 3.84LGVLMFAKK881 pKa = 10.62ANLEE885 pKa = 3.95LSDD888 pKa = 4.14FTWSCAVIGNKK899 pKa = 9.78KK900 pKa = 10.19VDD902 pKa = 3.8VKK904 pKa = 11.15KK905 pKa = 10.73CGYY908 pKa = 8.68DD909 pKa = 3.07TFHH912 pKa = 7.33QYY914 pKa = 10.06IGLEE918 pKa = 4.29PISDD922 pKa = 3.92YY923 pKa = 10.98FADD926 pKa = 5.01EE927 pKa = 4.41IDD929 pKa = 4.31DD930 pKa = 3.46GTMFVKK936 pKa = 10.42KK937 pKa = 10.05DD938 pKa = 3.65FKK940 pKa = 10.53IGKK943 pKa = 9.2INLIVDD949 pKa = 4.46LPSEE953 pKa = 4.52LFQTIAKK960 pKa = 8.25KK961 pKa = 10.48PKK963 pKa = 7.99VAMVASACKK972 pKa = 10.39GCLSCSSGINCDD984 pKa = 4.28LEE986 pKa = 4.43FTSDD990 pKa = 3.84SIFSCRR996 pKa = 11.84ISWEE1000 pKa = 4.12HH1001 pKa = 6.14CTSEE1005 pKa = 4.19PEE1007 pKa = 3.91QIAMKK1012 pKa = 10.47KK1013 pKa = 7.53GTNVFKK1019 pKa = 11.07LKK1021 pKa = 10.39PCFALKK1027 pKa = 10.15IQLLQKK1033 pKa = 10.34IILVPSDD1040 pKa = 4.14DD1041 pKa = 3.52EE1042 pKa = 5.24SMALEE1047 pKa = 4.21FEE1049 pKa = 4.59TKK1051 pKa = 10.15NVEE1054 pKa = 3.68ISDD1057 pKa = 4.01PEE1059 pKa = 4.3TIIDD1063 pKa = 4.07HH1064 pKa = 6.9NDD1066 pKa = 3.07DD1067 pKa = 3.68AFNEE1071 pKa = 4.19EE1072 pKa = 4.09VEE1074 pKa = 4.3YY1075 pKa = 11.1DD1076 pKa = 3.2SDD1078 pKa = 3.9TSFKK1082 pKa = 11.11SVWDD1086 pKa = 4.14YY1087 pKa = 11.46IKK1089 pKa = 11.26SPFNWVASFFGDD1101 pKa = 3.39FFEE1104 pKa = 4.37IVRR1107 pKa = 11.84VLLVLTCCAVLFISISKK1124 pKa = 10.18LYY1126 pKa = 8.94EE1127 pKa = 3.77ICHH1130 pKa = 6.35EE1131 pKa = 4.42YY1132 pKa = 10.83YY1133 pKa = 10.69SQEE1136 pKa = 3.98QYY1138 pKa = 10.99KK1139 pKa = 10.42KK1140 pKa = 10.49SSRR1143 pKa = 11.84KK1144 pKa = 9.6KK1145 pKa = 10.66EE1146 pKa = 3.74GDD1148 pKa = 4.18LEE1150 pKa = 6.33DD1151 pKa = 5.81DD1152 pKa = 4.71DD1153 pKa = 4.91EE1154 pKa = 6.36KK1155 pKa = 11.17KK1156 pKa = 10.97AQDD1159 pKa = 4.98DD1160 pKa = 4.13IIKK1163 pKa = 10.32SLRR1166 pKa = 11.84NQVTEE1171 pKa = 3.65DD1172 pKa = 3.09TTYY1175 pKa = 11.1KK1176 pKa = 10.53RR1177 pKa = 11.84NSNKK1181 pKa = 8.24STRR1184 pKa = 11.84KK1185 pKa = 10.15APFDD1189 pKa = 3.75DD1190 pKa = 5.22LIIRR1194 pKa = 11.84VV1195 pKa = 3.71
MM1 pKa = 7.91RR2 pKa = 11.84IKK4 pKa = 10.86DD5 pKa = 3.61FFVIMLLFGLNLVFLSIFINASIRR29 pKa = 11.84GDD31 pKa = 3.46HH32 pKa = 6.39EE33 pKa = 4.65VSQEE37 pKa = 3.89TSFEE41 pKa = 4.11EE42 pKa = 4.07LSKK45 pKa = 11.17EE46 pKa = 3.91VMEE49 pKa = 4.29AHH51 pKa = 6.03EE52 pKa = 4.09QGKK55 pKa = 9.67IGKK58 pKa = 8.54SLVSSRR64 pKa = 11.84VEE66 pKa = 3.64AFEE69 pKa = 4.45SFNKK73 pKa = 10.15EE74 pKa = 3.73PIEE77 pKa = 3.94TSRR80 pKa = 11.84RR81 pKa = 11.84PEE83 pKa = 3.67MNDD86 pKa = 4.02RR87 pKa = 11.84ITQTKK92 pKa = 9.63PGIDD96 pKa = 3.43DD97 pKa = 3.44QMIPKK102 pKa = 9.76HH103 pKa = 6.25GEE105 pKa = 4.16GKK107 pKa = 10.16RR108 pKa = 11.84EE109 pKa = 3.94APPPSKK115 pKa = 11.0VDD117 pKa = 3.74LSCANINPEE126 pKa = 3.68NCKK129 pKa = 10.34VVGKK133 pKa = 8.49TPFNFYY139 pKa = 10.59YY140 pKa = 10.31QISDD144 pKa = 3.68SEE146 pKa = 4.33EE147 pKa = 4.25TISCLSSKK155 pKa = 10.82VEE157 pKa = 4.57DD158 pKa = 4.35LKK160 pKa = 11.51ACNPQSMHH168 pKa = 5.85VFTSAPVLPITTILNKK184 pKa = 10.04RR185 pKa = 11.84VLSVGSKK192 pKa = 10.64YY193 pKa = 10.83YY194 pKa = 9.78IQNSIDD200 pKa = 3.44SSSFPVQDD208 pKa = 3.37SGNVQLGLVNIYY220 pKa = 8.46SVRR223 pKa = 11.84LTGEE227 pKa = 3.98CNITKK232 pKa = 10.03VSLTNPFFVTLTSSNKK248 pKa = 6.99EE249 pKa = 3.8TFYY252 pKa = 10.94SIKK255 pKa = 10.48KK256 pKa = 9.94IGADD260 pKa = 3.53DD261 pKa = 3.72VNLIKK266 pKa = 10.76FSGEE270 pKa = 3.81KK271 pKa = 9.39TVEE274 pKa = 4.07LPSDD278 pKa = 3.87SVDD281 pKa = 3.43GNHH284 pKa = 6.49VLLCGDD290 pKa = 4.17KK291 pKa = 10.82VSEE294 pKa = 4.15IPKK297 pKa = 9.43IDD299 pKa = 3.56VNKK302 pKa = 10.28RR303 pKa = 11.84NCLVKK308 pKa = 10.93YY309 pKa = 9.93KK310 pKa = 10.54GKK312 pKa = 10.46RR313 pKa = 11.84YY314 pKa = 8.06QQSACVHH321 pKa = 5.66FNILRR326 pKa = 11.84YY327 pKa = 9.69LLVSIILFVPVSWFLNKK344 pKa = 9.97TKK346 pKa = 11.0DD347 pKa = 5.21SMFLWYY353 pKa = 10.0DD354 pKa = 3.01FLGIIFYY361 pKa = 9.9PILYY365 pKa = 9.1ILNWLWRR372 pKa = 11.84YY373 pKa = 9.47FPMKK377 pKa = 10.32CSNCGNFSLITHH389 pKa = 6.32SCYY392 pKa = 9.73DD393 pKa = 3.3TCVCNKK399 pKa = 9.9SKK401 pKa = 10.93AKK403 pKa = 10.35SEE405 pKa = 4.03HH406 pKa = 6.74AEE408 pKa = 3.75NCPIITGKK416 pKa = 10.48AFSKK420 pKa = 11.07SEE422 pKa = 3.97DD423 pKa = 3.45QDD425 pKa = 3.7VDD427 pKa = 3.82DD428 pKa = 5.73LSVEE432 pKa = 4.24EE433 pKa = 4.69TEE435 pKa = 5.08SLTGSKK441 pKa = 10.06KK442 pKa = 9.79PKK444 pKa = 9.23KK445 pKa = 9.86ASSKK449 pKa = 10.05FRR451 pKa = 11.84TFQLVVNTKK460 pKa = 9.66ISTGFLMFVTKK471 pKa = 10.65VLLGFLIFSQMPKK484 pKa = 9.75TMASKK489 pKa = 8.98TATQTISQTEE499 pKa = 4.24TQVLCVSYY507 pKa = 10.81CSAKK511 pKa = 10.06PGCNKK516 pKa = 9.72FLFKK520 pKa = 11.1NEE522 pKa = 4.06EE523 pKa = 3.96VCIDD527 pKa = 3.85NPKK530 pKa = 10.34LKK532 pKa = 10.31CNCLIRR538 pKa = 11.84DD539 pKa = 3.55GMINEE544 pKa = 4.01EE545 pKa = 4.22AYY547 pKa = 10.03INGTRR552 pKa = 11.84YY553 pKa = 9.72SVSMVDD559 pKa = 2.89NCLDD563 pKa = 3.54RR564 pKa = 11.84RR565 pKa = 11.84CEE567 pKa = 3.76FSSNKK572 pKa = 9.48VEE574 pKa = 5.25DD575 pKa = 4.26LLACRR580 pKa = 11.84LGCGQLKK587 pKa = 10.08KK588 pKa = 10.74LKK590 pKa = 10.26SSKK593 pKa = 10.26LQSKK597 pKa = 9.54ALKK600 pKa = 10.08QGIVPRR606 pKa = 11.84GASISQIYY614 pKa = 10.31SSLRR618 pKa = 11.84LQDD621 pKa = 4.11GYY623 pKa = 10.78IDD625 pKa = 4.09SEE627 pKa = 4.45EE628 pKa = 4.29SLHH631 pKa = 7.34SLGDD635 pKa = 3.6LPEE638 pKa = 4.31NKK640 pKa = 9.71VKK642 pKa = 10.56IEE644 pKa = 4.67DD645 pKa = 3.48EE646 pKa = 4.84DD647 pKa = 3.86IPEE650 pKa = 4.44GVLPRR655 pKa = 11.84QSFVYY660 pKa = 10.59NSIVDD665 pKa = 3.57GKK667 pKa = 9.86YY668 pKa = 9.85RR669 pKa = 11.84YY670 pKa = 9.25LISMDD675 pKa = 4.97AIQSTGYY682 pKa = 9.78VYY684 pKa = 11.43SMNDD688 pKa = 2.79AKK690 pKa = 10.31MAPPQEE696 pKa = 3.72LVVFVKK702 pKa = 10.49EE703 pKa = 4.04AGVTYY708 pKa = 9.9TMKK711 pKa = 10.01TLYY714 pKa = 8.81YY715 pKa = 8.02TAPISATHH723 pKa = 4.55THH725 pKa = 6.12VYY727 pKa = 7.95STCTGNCEE735 pKa = 4.35TCRR738 pKa = 11.84KK739 pKa = 7.05EE740 pKa = 4.37HH741 pKa = 7.07PISGYY746 pKa = 8.75QDD748 pKa = 3.24YY749 pKa = 11.11CIAPTSNWGCEE760 pKa = 4.03EE761 pKa = 4.63LGCLAIGEE769 pKa = 4.64GATCGYY775 pKa = 9.0CRR777 pKa = 11.84NVYY780 pKa = 10.1DD781 pKa = 4.27LSKK784 pKa = 10.35KK785 pKa = 9.69YY786 pKa = 10.14SVRR789 pKa = 11.84QILTSHH795 pKa = 5.46VTVKK799 pKa = 10.58VCFKK803 pKa = 10.59GFAGTGCQTITDD815 pKa = 4.06SVPFQNSYY823 pKa = 10.06YY824 pKa = 10.03QLSIDD829 pKa = 4.63ADD831 pKa = 3.84LHH833 pKa = 7.16NDD835 pKa = 3.45DD836 pKa = 4.57LTAGSRR842 pKa = 11.84VAVDD846 pKa = 3.5PNGMLFKK853 pKa = 11.33GNIANLKK860 pKa = 10.15DD861 pKa = 3.19SSLAFGHH868 pKa = 6.23PQLNEE873 pKa = 3.84LGVLMFAKK881 pKa = 10.62ANLEE885 pKa = 3.95LSDD888 pKa = 4.14FTWSCAVIGNKK899 pKa = 9.78KK900 pKa = 10.19VDD902 pKa = 3.8VKK904 pKa = 11.15KK905 pKa = 10.73CGYY908 pKa = 8.68DD909 pKa = 3.07TFHH912 pKa = 7.33QYY914 pKa = 10.06IGLEE918 pKa = 4.29PISDD922 pKa = 3.92YY923 pKa = 10.98FADD926 pKa = 5.01EE927 pKa = 4.41IDD929 pKa = 4.31DD930 pKa = 3.46GTMFVKK936 pKa = 10.42KK937 pKa = 10.05DD938 pKa = 3.65FKK940 pKa = 10.53IGKK943 pKa = 9.2INLIVDD949 pKa = 4.46LPSEE953 pKa = 4.52LFQTIAKK960 pKa = 8.25KK961 pKa = 10.48PKK963 pKa = 7.99VAMVASACKK972 pKa = 10.39GCLSCSSGINCDD984 pKa = 4.28LEE986 pKa = 4.43FTSDD990 pKa = 3.84SIFSCRR996 pKa = 11.84ISWEE1000 pKa = 4.12HH1001 pKa = 6.14CTSEE1005 pKa = 4.19PEE1007 pKa = 3.91QIAMKK1012 pKa = 10.47KK1013 pKa = 7.53GTNVFKK1019 pKa = 11.07LKK1021 pKa = 10.39PCFALKK1027 pKa = 10.15IQLLQKK1033 pKa = 10.34IILVPSDD1040 pKa = 4.14DD1041 pKa = 3.52EE1042 pKa = 5.24SMALEE1047 pKa = 4.21FEE1049 pKa = 4.59TKK1051 pKa = 10.15NVEE1054 pKa = 3.68ISDD1057 pKa = 4.01PEE1059 pKa = 4.3TIIDD1063 pKa = 4.07HH1064 pKa = 6.9NDD1066 pKa = 3.07DD1067 pKa = 3.68AFNEE1071 pKa = 4.19EE1072 pKa = 4.09VEE1074 pKa = 4.3YY1075 pKa = 11.1DD1076 pKa = 3.2SDD1078 pKa = 3.9TSFKK1082 pKa = 11.11SVWDD1086 pKa = 4.14YY1087 pKa = 11.46IKK1089 pKa = 11.26SPFNWVASFFGDD1101 pKa = 3.39FFEE1104 pKa = 4.37IVRR1107 pKa = 11.84VLLVLTCCAVLFISISKK1124 pKa = 10.18LYY1126 pKa = 8.94EE1127 pKa = 3.77ICHH1130 pKa = 6.35EE1131 pKa = 4.42YY1132 pKa = 10.83YY1133 pKa = 10.69SQEE1136 pKa = 3.98QYY1138 pKa = 10.99KK1139 pKa = 10.42KK1140 pKa = 10.49SSRR1143 pKa = 11.84KK1144 pKa = 9.6KK1145 pKa = 10.66EE1146 pKa = 3.74GDD1148 pKa = 4.18LEE1150 pKa = 6.33DD1151 pKa = 5.81DD1152 pKa = 4.71DD1153 pKa = 4.91EE1154 pKa = 6.36KK1155 pKa = 11.17KK1156 pKa = 10.97AQDD1159 pKa = 4.98DD1160 pKa = 4.13IIKK1163 pKa = 10.32SLRR1166 pKa = 11.84NQVTEE1171 pKa = 3.65DD1172 pKa = 3.09TTYY1175 pKa = 11.1KK1176 pKa = 10.53RR1177 pKa = 11.84NSNKK1181 pKa = 8.24STRR1184 pKa = 11.84KK1185 pKa = 10.15APFDD1189 pKa = 3.75DD1190 pKa = 5.22LIIRR1194 pKa = 11.84VV1195 pKa = 3.71
Molecular weight: 134.32 kDa
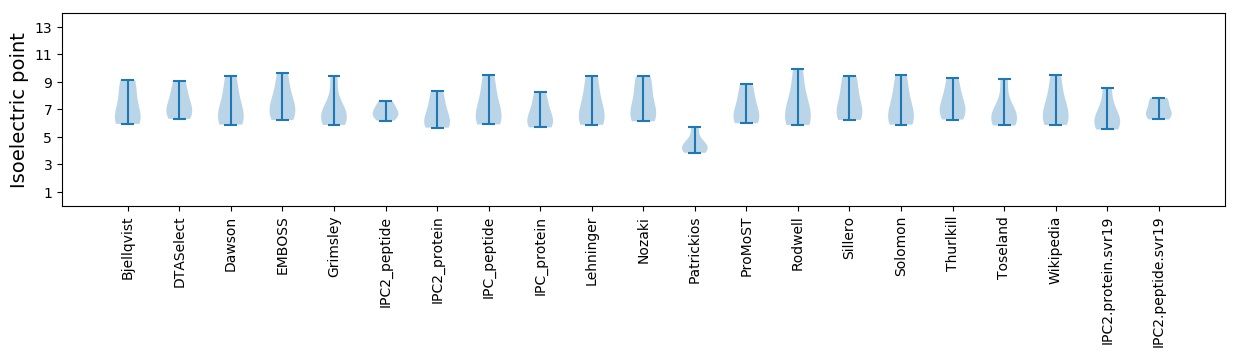
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2Y3N7|F2Y3N7_9VIRU Nucleocapsid protein OS=Soybean vein necrosis virus OX=980895 PE=3 SV=1
MM1 pKa = 7.51PQTAGPSNAKK11 pKa = 8.57PAKK14 pKa = 8.76ITEE17 pKa = 4.3SNLAKK22 pKa = 10.46LLKK25 pKa = 10.25FEE27 pKa = 4.68EE28 pKa = 5.13DD29 pKa = 3.57IEE31 pKa = 4.35FEE33 pKa = 4.3KK34 pKa = 11.19NSTGFKK40 pKa = 10.25FSEE43 pKa = 5.09FYY45 pKa = 8.4KK46 pKa = 9.31THH48 pKa = 6.22MGRR51 pKa = 11.84KK52 pKa = 8.72FRR54 pKa = 11.84YY55 pKa = 9.21ASALTFLKK63 pKa = 10.41NRR65 pKa = 11.84KK66 pKa = 9.6AIVNMCKK73 pKa = 10.24KK74 pKa = 9.82GTFNFDD80 pKa = 2.95GQTVKK85 pKa = 10.99LSVEE89 pKa = 4.25SGDD92 pKa = 4.25DD93 pKa = 3.3NSFTFKK99 pKa = 10.92RR100 pKa = 11.84LDD102 pKa = 3.09SFLRR106 pKa = 11.84VKK108 pKa = 10.19MLEE111 pKa = 3.95HH112 pKa = 6.46NFAVFDD118 pKa = 4.0GTNEE122 pKa = 3.77EE123 pKa = 4.71AKK125 pKa = 10.55QSLCNDD131 pKa = 4.09LATIPLVQAYY141 pKa = 9.83GLTVKK146 pKa = 10.8DD147 pKa = 3.56KK148 pKa = 10.61MSAKK152 pKa = 9.6LAIMIGGSLPLLASITGCEE171 pKa = 4.16AYY173 pKa = 10.67CFGLAIFQDD182 pKa = 3.99LKK184 pKa = 11.05KK185 pKa = 9.96EE186 pKa = 3.77QLGIVNFDD194 pKa = 3.66TKK196 pKa = 11.15AQAAKK201 pKa = 10.03VASVLDD207 pKa = 3.64AKK209 pKa = 10.71GFKK212 pKa = 9.03FTEE215 pKa = 4.54EE216 pKa = 3.95KK217 pKa = 10.84NQTLRR222 pKa = 11.84LIAEE226 pKa = 4.42ILKK229 pKa = 11.01DD230 pKa = 3.41MAPQMRR236 pKa = 11.84GVASLEE242 pKa = 4.21KK243 pKa = 10.21YY244 pKa = 10.27NEE246 pKa = 4.03QIGIISDD253 pKa = 3.93IIGVHH258 pKa = 6.18FEE260 pKa = 4.12MPGKK264 pKa = 10.31KK265 pKa = 9.66DD266 pKa = 3.53GKK268 pKa = 9.79GKK270 pKa = 10.11KK271 pKa = 9.74SKK273 pKa = 10.26EE274 pKa = 3.77FSVV277 pKa = 3.57
MM1 pKa = 7.51PQTAGPSNAKK11 pKa = 8.57PAKK14 pKa = 8.76ITEE17 pKa = 4.3SNLAKK22 pKa = 10.46LLKK25 pKa = 10.25FEE27 pKa = 4.68EE28 pKa = 5.13DD29 pKa = 3.57IEE31 pKa = 4.35FEE33 pKa = 4.3KK34 pKa = 11.19NSTGFKK40 pKa = 10.25FSEE43 pKa = 5.09FYY45 pKa = 8.4KK46 pKa = 9.31THH48 pKa = 6.22MGRR51 pKa = 11.84KK52 pKa = 8.72FRR54 pKa = 11.84YY55 pKa = 9.21ASALTFLKK63 pKa = 10.41NRR65 pKa = 11.84KK66 pKa = 9.6AIVNMCKK73 pKa = 10.24KK74 pKa = 9.82GTFNFDD80 pKa = 2.95GQTVKK85 pKa = 10.99LSVEE89 pKa = 4.25SGDD92 pKa = 4.25DD93 pKa = 3.3NSFTFKK99 pKa = 10.92RR100 pKa = 11.84LDD102 pKa = 3.09SFLRR106 pKa = 11.84VKK108 pKa = 10.19MLEE111 pKa = 3.95HH112 pKa = 6.46NFAVFDD118 pKa = 4.0GTNEE122 pKa = 3.77EE123 pKa = 4.71AKK125 pKa = 10.55QSLCNDD131 pKa = 4.09LATIPLVQAYY141 pKa = 9.83GLTVKK146 pKa = 10.8DD147 pKa = 3.56KK148 pKa = 10.61MSAKK152 pKa = 9.6LAIMIGGSLPLLASITGCEE171 pKa = 4.16AYY173 pKa = 10.67CFGLAIFQDD182 pKa = 3.99LKK184 pKa = 11.05KK185 pKa = 9.96EE186 pKa = 3.77QLGIVNFDD194 pKa = 3.66TKK196 pKa = 11.15AQAAKK201 pKa = 10.03VASVLDD207 pKa = 3.64AKK209 pKa = 10.71GFKK212 pKa = 9.03FTEE215 pKa = 4.54EE216 pKa = 3.95KK217 pKa = 10.84NQTLRR222 pKa = 11.84LIAEE226 pKa = 4.42ILKK229 pKa = 11.01DD230 pKa = 3.41MAPQMRR236 pKa = 11.84GVASLEE242 pKa = 4.21KK243 pKa = 10.21YY244 pKa = 10.27NEE246 pKa = 4.03QIGIISDD253 pKa = 3.93IIGVHH258 pKa = 6.18FEE260 pKa = 4.12MPGKK264 pKa = 10.31KK265 pKa = 9.66DD266 pKa = 3.53GKK268 pKa = 9.79GKK270 pKa = 10.11KK271 pKa = 9.74SKK273 pKa = 10.26EE274 pKa = 3.77FSVV277 pKa = 3.57
Molecular weight: 30.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5159 |
277 |
2931 |
1031.8 |
117.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.09 ± 0.677 | 2.384 ± 0.5 |
5.99 ± 0.328 | 6.978 ± 0.395 |
5.02 ± 0.508 | 5.117 ± 0.667 |
1.667 ± 0.209 | 7.269 ± 0.237 |
9.304 ± 0.545 | 9.033 ± 0.666 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.198 ± 0.451 | 5.582 ± 0.341 |
3.024 ± 0.422 | 2.811 ± 0.265 |
3.353 ± 0.391 | 9.343 ± 0.324 |
5.679 ± 0.5 | 5.408 ± 0.5 |
0.678 ± 0.165 | 4.071 ± 0.546 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
