
Sodalis praecaptivus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Bruguierivoracaceae; Sodalis
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
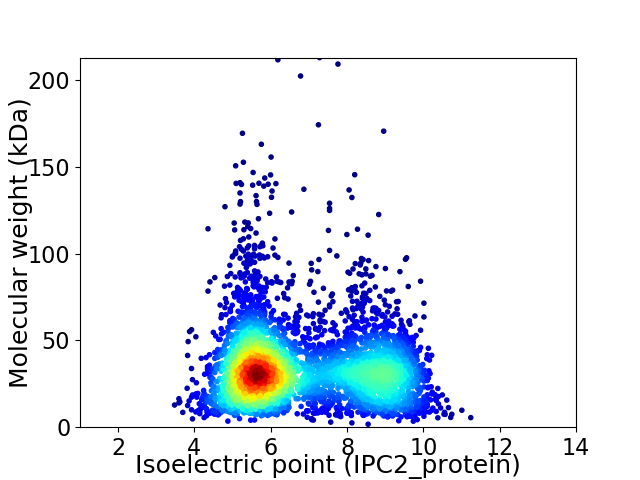
Virtual 2D-PAGE plot for 4281 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
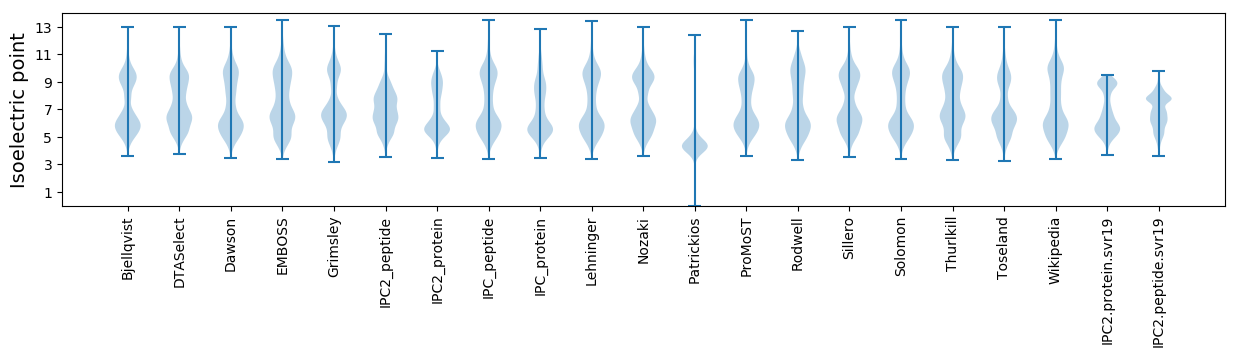
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W0HPK6|W0HPK6_9GAMM Flagellar secretion chaperone FliS OS=Sodalis praecaptivus OX=1239307 GN=fliS2 PE=3 SV=1
MM1 pKa = 7.53GFSQAVSGLNAASSNLDD18 pKa = 3.36VIGNNIANAEE28 pKa = 4.12TSGFKK33 pKa = 10.4SGSVSFADD41 pKa = 3.57VFSSSKK47 pKa = 10.56VGLGVKK53 pKa = 9.34VASVTQNFNDD63 pKa = 4.16GTVSTTDD70 pKa = 2.89NGLDD74 pKa = 3.37VAISNNGFFRR84 pKa = 11.84LVDD87 pKa = 3.74GSGAVYY93 pKa = 10.16YY94 pKa = 10.67SRR96 pKa = 11.84NGEE99 pKa = 3.83FSLNADD105 pKa = 3.74RR106 pKa = 11.84QLVNSSGYY114 pKa = 10.47VVTGYY119 pKa = 10.53AASGNPATIQQGADD133 pKa = 3.44PVALTIPSQGLSASATTSGSMVMSLTSSATSVGTTAFDD171 pKa = 4.63PNDD174 pKa = 3.21SSTYY178 pKa = 10.71SFSQPLTTYY187 pKa = 11.0DD188 pKa = 3.99SLGNPHH194 pKa = 6.48NVSLYY199 pKa = 9.62FAKK202 pKa = 10.22TEE204 pKa = 4.33DD205 pKa = 4.03NTWEE209 pKa = 4.26VYY211 pKa = 10.62SVDD214 pKa = 3.4NSNGDD219 pKa = 3.37GTVNDD224 pKa = 3.71VGTLNFDD231 pKa = 3.76TNGQLTGTDD240 pKa = 3.08TFSINMAALNGAPAQTFNLSFEE262 pKa = 4.51GTRR265 pKa = 11.84QQYY268 pKa = 10.8ASSSSVSSQTQDD280 pKa = 2.74GYY282 pKa = 11.5SAGDD286 pKa = 3.47LTSYY290 pKa = 9.52TINDD294 pKa = 4.22DD295 pKa = 3.62GTISGTYY302 pKa = 10.51SNGKK306 pKa = 6.7TQLLGQIVLANFANPQGLQSDD327 pKa = 4.63GNNLWSATSASGQAIVGTAGSGTFGTLTSGALEE360 pKa = 4.15SSNVDD365 pKa = 3.63LSQEE369 pKa = 4.1LVNMIVAQRR378 pKa = 11.84NYY380 pKa = 10.23QSNAQTIKK388 pKa = 9.16TQDD391 pKa = 4.02AILQTLVSLRR401 pKa = 3.71
MM1 pKa = 7.53GFSQAVSGLNAASSNLDD18 pKa = 3.36VIGNNIANAEE28 pKa = 4.12TSGFKK33 pKa = 10.4SGSVSFADD41 pKa = 3.57VFSSSKK47 pKa = 10.56VGLGVKK53 pKa = 9.34VASVTQNFNDD63 pKa = 4.16GTVSTTDD70 pKa = 2.89NGLDD74 pKa = 3.37VAISNNGFFRR84 pKa = 11.84LVDD87 pKa = 3.74GSGAVYY93 pKa = 10.16YY94 pKa = 10.67SRR96 pKa = 11.84NGEE99 pKa = 3.83FSLNADD105 pKa = 3.74RR106 pKa = 11.84QLVNSSGYY114 pKa = 10.47VVTGYY119 pKa = 10.53AASGNPATIQQGADD133 pKa = 3.44PVALTIPSQGLSASATTSGSMVMSLTSSATSVGTTAFDD171 pKa = 4.63PNDD174 pKa = 3.21SSTYY178 pKa = 10.71SFSQPLTTYY187 pKa = 11.0DD188 pKa = 3.99SLGNPHH194 pKa = 6.48NVSLYY199 pKa = 9.62FAKK202 pKa = 10.22TEE204 pKa = 4.33DD205 pKa = 4.03NTWEE209 pKa = 4.26VYY211 pKa = 10.62SVDD214 pKa = 3.4NSNGDD219 pKa = 3.37GTVNDD224 pKa = 3.71VGTLNFDD231 pKa = 3.76TNGQLTGTDD240 pKa = 3.08TFSINMAALNGAPAQTFNLSFEE262 pKa = 4.51GTRR265 pKa = 11.84QQYY268 pKa = 10.8ASSSSVSSQTQDD280 pKa = 2.74GYY282 pKa = 11.5SAGDD286 pKa = 3.47LTSYY290 pKa = 9.52TINDD294 pKa = 4.22DD295 pKa = 3.62GTISGTYY302 pKa = 10.51SNGKK306 pKa = 6.7TQLLGQIVLANFANPQGLQSDD327 pKa = 4.63GNNLWSATSASGQAIVGTAGSGTFGTLTSGALEE360 pKa = 4.15SSNVDD365 pKa = 3.63LSQEE369 pKa = 4.1LVNMIVAQRR378 pKa = 11.84NYY380 pKa = 10.23QSNAQTIKK388 pKa = 9.16TQDD391 pKa = 4.02AILQTLVSLRR401 pKa = 3.71
Molecular weight: 41.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W0HRQ2|W0HRQ2_9GAMM 2Fe-2S ferredoxin OS=Sodalis praecaptivus OX=1239307 GN=yfaE PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.68RR12 pKa = 11.84NRR14 pKa = 11.84THH16 pKa = 7.28GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.36GRR39 pKa = 11.84TRR41 pKa = 11.84LTVSSKK47 pKa = 11.13
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.68RR12 pKa = 11.84NRR14 pKa = 11.84THH16 pKa = 7.28GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.36GRR39 pKa = 11.84TRR41 pKa = 11.84LTVSSKK47 pKa = 11.13
Molecular weight: 5.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1400054 |
16 |
1965 |
327.0 |
35.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.129 ± 0.045 | 1.091 ± 0.012 |
5.195 ± 0.027 | 5.013 ± 0.036 |
3.639 ± 0.024 | 7.703 ± 0.035 |
2.371 ± 0.017 | 5.47 ± 0.028 |
3.341 ± 0.03 | 11.356 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.574 ± 0.016 | 3.26 ± 0.025 |
4.912 ± 0.032 | 4.517 ± 0.032 |
6.531 ± 0.034 | 5.648 ± 0.025 |
5.228 ± 0.023 | 6.886 ± 0.031 |
1.4 ± 0.016 | 2.737 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
