
Beihai sobemo-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.52
Get precalculated fractions of proteins
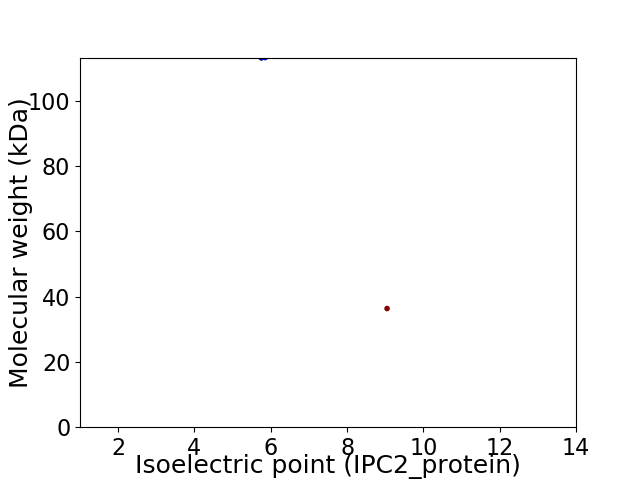
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEA9|A0A1L3KEA9_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 9 OX=1922706 PE=4 SV=1
MM1 pKa = 7.61AVTIEE6 pKa = 4.26ITDD9 pKa = 3.68DD10 pKa = 3.73MIAQAVLYY18 pKa = 9.37SKK20 pKa = 11.23YY21 pKa = 10.05GGAVGAVMIAIRR33 pKa = 11.84MGLLRR38 pKa = 11.84FLVGVVYY45 pKa = 10.85NVFGFLGFAATTSVGWWRR63 pKa = 11.84YY64 pKa = 8.88HH65 pKa = 4.63SQIGAKK71 pKa = 9.34EE72 pKa = 3.79LAVCKK77 pKa = 9.98TGFVTYY83 pKa = 10.63GDD85 pKa = 3.37RR86 pKa = 11.84TFYY89 pKa = 10.67RR90 pKa = 11.84DD91 pKa = 3.26PSDD94 pKa = 3.48LVLYY98 pKa = 9.61SVKK101 pKa = 10.16PGRR104 pKa = 11.84ARR106 pKa = 11.84FVEE109 pKa = 4.59GQFVLSDD116 pKa = 3.56YY117 pKa = 11.35VLVPEE122 pKa = 4.32EE123 pKa = 4.25TGTIAEE129 pKa = 4.28SALYY133 pKa = 10.38GSDD136 pKa = 4.15PIATRR141 pKa = 11.84SVAPNDD147 pKa = 3.58YY148 pKa = 8.9VSVYY152 pKa = 10.08VVHH155 pKa = 7.81DD156 pKa = 4.16DD157 pKa = 4.02GYY159 pKa = 10.25HH160 pKa = 5.04YY161 pKa = 10.9LGGGFRR167 pKa = 11.84EE168 pKa = 4.19AAFLITAAHH177 pKa = 7.61AITDD181 pKa = 4.14SLDD184 pKa = 3.13HH185 pKa = 7.16RR186 pKa = 11.84IALSKK191 pKa = 10.63DD192 pKa = 2.92GKK194 pKa = 10.67KK195 pKa = 10.02FYY197 pKa = 10.83HH198 pKa = 6.26PGKK201 pKa = 9.95VIRR204 pKa = 11.84HH205 pKa = 5.78FVNDD209 pKa = 3.9DD210 pKa = 3.41YY211 pKa = 11.99VRR213 pKa = 11.84CTGDD217 pKa = 3.23DD218 pKa = 3.15VGAFEE223 pKa = 6.05LSAADD228 pKa = 3.56WAVLGVRR235 pKa = 11.84SVKK238 pKa = 10.2PSVYY242 pKa = 10.02SATGMSRR249 pKa = 11.84IEE251 pKa = 4.08VFGRR255 pKa = 11.84DD256 pKa = 3.31PVGVLKK262 pKa = 11.04AGVGNLLPPTQKK274 pKa = 10.21QKK276 pKa = 10.55YY277 pKa = 8.38LGVVPHH283 pKa = 6.58SASTMKK289 pKa = 10.41GFSGSPVYY297 pKa = 10.37TIGEE301 pKa = 4.25TGRR304 pKa = 11.84KK305 pKa = 8.73IVGLHH310 pKa = 5.34IAGGTGTEE318 pKa = 3.8NYY320 pKa = 7.1MASVHH325 pKa = 6.56EE326 pKa = 4.05IHH328 pKa = 6.49QLRR331 pKa = 11.84RR332 pKa = 11.84KK333 pKa = 9.93LGLIPEE339 pKa = 4.35VPMVAEE345 pKa = 4.97ASPDD349 pKa = 3.32MKK351 pKa = 10.45HH352 pKa = 6.02RR353 pKa = 11.84QFDD356 pKa = 3.56RR357 pKa = 11.84HH358 pKa = 4.99EE359 pKa = 5.07HH360 pKa = 5.99NDD362 pKa = 3.48LMDD365 pKa = 4.7DD366 pKa = 4.15LSRR369 pKa = 11.84VSKK372 pKa = 10.77HH373 pKa = 5.63GGLLRR378 pKa = 11.84VYY380 pKa = 10.95DD381 pKa = 4.36DD382 pKa = 4.91LAGNDD387 pKa = 3.77YY388 pKa = 11.27AEE390 pKa = 4.66SADD393 pKa = 3.39IPKK396 pKa = 10.64AVGTPVTGLEE406 pKa = 4.4PEE408 pKa = 4.25VSSGSGADD416 pKa = 3.91LVPALAPVPSTGNASAKK433 pKa = 9.66EE434 pKa = 3.97KK435 pKa = 10.49AASEE439 pKa = 4.32VADD442 pKa = 3.61VSTDD446 pKa = 3.13ADD448 pKa = 3.89LSHH451 pKa = 7.44PDD453 pKa = 3.79EE454 pKa = 5.16VPDD457 pKa = 3.96EE458 pKa = 4.44LDD460 pKa = 3.3FSKK463 pKa = 10.82LSEE466 pKa = 4.43EE467 pKa = 4.41EE468 pKa = 4.03PVPPIGVKK476 pKa = 9.82RR477 pKa = 11.84VRR479 pKa = 11.84NKK481 pKa = 10.28KK482 pKa = 9.91VGGIGAFLGTTALAGAVIGKK502 pKa = 9.46VGSAFDD508 pKa = 4.77PKK510 pKa = 10.59WLDD513 pKa = 3.28CTAASLAEE521 pKa = 4.1ASAASYY527 pKa = 10.55HH528 pKa = 7.16AMLNSPNFDD537 pKa = 3.19RR538 pKa = 11.84YY539 pKa = 10.04RR540 pKa = 11.84EE541 pKa = 3.94YY542 pKa = 10.32MSASGVVAADD552 pKa = 3.44QQRR555 pKa = 11.84TMKK558 pKa = 10.75GVDD561 pKa = 3.31GTDD564 pKa = 3.24LARR567 pKa = 11.84LFGTGVSNRR576 pKa = 11.84SRR578 pKa = 11.84KK579 pKa = 9.3KK580 pKa = 9.73ALKK583 pKa = 9.81RR584 pKa = 11.84LPARR588 pKa = 11.84FLEE591 pKa = 4.5VVEE594 pKa = 4.65KK595 pKa = 11.05LGLPVKK601 pKa = 10.48QYY603 pKa = 11.15AQWTLPPAGVEE614 pKa = 4.07AMEE617 pKa = 5.0HH618 pKa = 5.91SLATQLGRR626 pKa = 11.84TCSSCWPEE634 pKa = 3.5EE635 pKa = 3.89TRR637 pKa = 11.84RR638 pKa = 11.84QFANKK643 pKa = 9.93SGPQYY648 pKa = 10.89DD649 pKa = 3.33IFLEE653 pKa = 4.54EE654 pKa = 4.11VARR657 pKa = 11.84YY658 pKa = 6.92PANRR662 pKa = 11.84ADD664 pKa = 3.54AFANVHH670 pKa = 5.71SKK672 pKa = 7.45VTRR675 pKa = 11.84FVLGLDD681 pKa = 3.78GDD683 pKa = 4.54KK684 pKa = 11.1SAGWSQHH691 pKa = 4.84FRR693 pKa = 11.84PGTKK697 pKa = 9.77RR698 pKa = 11.84SWQDD702 pKa = 3.21TEE704 pKa = 4.36GLTLASYY711 pKa = 7.6LTRR714 pKa = 11.84CRR716 pKa = 11.84LLLRR720 pKa = 11.84AAVGPCVMAQMTPSQLIEE738 pKa = 4.86AGLSDD743 pKa = 4.74PRR745 pKa = 11.84TLFIKK750 pKa = 10.67NEE752 pKa = 3.83PHH754 pKa = 6.67SDD756 pKa = 3.3SKK758 pKa = 11.43VADD761 pKa = 3.84GRR763 pKa = 11.84WRR765 pKa = 11.84LIWGASLVDD774 pKa = 3.41VCVASVTCRR783 pKa = 11.84RR784 pKa = 11.84QDD786 pKa = 3.19KK787 pKa = 11.03LDD789 pKa = 3.34IEE791 pKa = 4.79QYY793 pKa = 10.35QGGPVEE799 pKa = 5.43GGHH802 pKa = 5.23QQAAGLGHH810 pKa = 7.13HH811 pKa = 7.27DD812 pKa = 3.56AGISRR817 pKa = 11.84LCVEE821 pKa = 4.85FQRR824 pKa = 11.84LIDD827 pKa = 3.54TGLQVFDD834 pKa = 4.76SDD836 pKa = 3.96ASGWDD841 pKa = 3.09MSVNRR846 pKa = 11.84DD847 pKa = 3.36SLYY850 pKa = 11.05ADD852 pKa = 3.63ALRR855 pKa = 11.84RR856 pKa = 11.84ILLYY860 pKa = 10.19EE861 pKa = 4.1GRR863 pKa = 11.84GKK865 pKa = 10.65AVFEE869 pKa = 3.98RR870 pKa = 11.84LALCEE875 pKa = 3.72AAANSAHH882 pKa = 5.64VVLIGGNLWEE892 pKa = 4.17VLKK895 pKa = 10.86PGITASGILSTTAQNSFIRR914 pKa = 11.84ALLYY918 pKa = 10.2AFVGVRR924 pKa = 11.84HH925 pKa = 5.84SVVAGDD931 pKa = 4.36DD932 pKa = 3.64GAGARR937 pKa = 11.84EE938 pKa = 3.92HH939 pKa = 7.04GYY941 pKa = 10.84DD942 pKa = 3.29HH943 pKa = 6.9VKK945 pKa = 10.75ALAEE949 pKa = 3.99YY950 pKa = 10.22GPEE953 pKa = 3.82EE954 pKa = 4.21KK955 pKa = 10.18QVNLYY960 pKa = 8.82TADD963 pKa = 3.5SGLEE967 pKa = 4.1FTSHH971 pKa = 5.68RR972 pKa = 11.84MRR974 pKa = 11.84RR975 pKa = 11.84GTDD978 pKa = 3.55GTWSAEE984 pKa = 3.57FLNLGKK990 pKa = 10.53ACARR994 pKa = 11.84LAFGDD999 pKa = 4.3TVTADD1004 pKa = 3.41QLAGLMFCVRR1014 pKa = 11.84NNPAEE1019 pKa = 4.33LKK1021 pKa = 9.44TLGDD1025 pKa = 3.56IAEE1028 pKa = 4.18EE1029 pKa = 4.12MGWPIAGVTPAFLPFLDD1046 pKa = 3.85
MM1 pKa = 7.61AVTIEE6 pKa = 4.26ITDD9 pKa = 3.68DD10 pKa = 3.73MIAQAVLYY18 pKa = 9.37SKK20 pKa = 11.23YY21 pKa = 10.05GGAVGAVMIAIRR33 pKa = 11.84MGLLRR38 pKa = 11.84FLVGVVYY45 pKa = 10.85NVFGFLGFAATTSVGWWRR63 pKa = 11.84YY64 pKa = 8.88HH65 pKa = 4.63SQIGAKK71 pKa = 9.34EE72 pKa = 3.79LAVCKK77 pKa = 9.98TGFVTYY83 pKa = 10.63GDD85 pKa = 3.37RR86 pKa = 11.84TFYY89 pKa = 10.67RR90 pKa = 11.84DD91 pKa = 3.26PSDD94 pKa = 3.48LVLYY98 pKa = 9.61SVKK101 pKa = 10.16PGRR104 pKa = 11.84ARR106 pKa = 11.84FVEE109 pKa = 4.59GQFVLSDD116 pKa = 3.56YY117 pKa = 11.35VLVPEE122 pKa = 4.32EE123 pKa = 4.25TGTIAEE129 pKa = 4.28SALYY133 pKa = 10.38GSDD136 pKa = 4.15PIATRR141 pKa = 11.84SVAPNDD147 pKa = 3.58YY148 pKa = 8.9VSVYY152 pKa = 10.08VVHH155 pKa = 7.81DD156 pKa = 4.16DD157 pKa = 4.02GYY159 pKa = 10.25HH160 pKa = 5.04YY161 pKa = 10.9LGGGFRR167 pKa = 11.84EE168 pKa = 4.19AAFLITAAHH177 pKa = 7.61AITDD181 pKa = 4.14SLDD184 pKa = 3.13HH185 pKa = 7.16RR186 pKa = 11.84IALSKK191 pKa = 10.63DD192 pKa = 2.92GKK194 pKa = 10.67KK195 pKa = 10.02FYY197 pKa = 10.83HH198 pKa = 6.26PGKK201 pKa = 9.95VIRR204 pKa = 11.84HH205 pKa = 5.78FVNDD209 pKa = 3.9DD210 pKa = 3.41YY211 pKa = 11.99VRR213 pKa = 11.84CTGDD217 pKa = 3.23DD218 pKa = 3.15VGAFEE223 pKa = 6.05LSAADD228 pKa = 3.56WAVLGVRR235 pKa = 11.84SVKK238 pKa = 10.2PSVYY242 pKa = 10.02SATGMSRR249 pKa = 11.84IEE251 pKa = 4.08VFGRR255 pKa = 11.84DD256 pKa = 3.31PVGVLKK262 pKa = 11.04AGVGNLLPPTQKK274 pKa = 10.21QKK276 pKa = 10.55YY277 pKa = 8.38LGVVPHH283 pKa = 6.58SASTMKK289 pKa = 10.41GFSGSPVYY297 pKa = 10.37TIGEE301 pKa = 4.25TGRR304 pKa = 11.84KK305 pKa = 8.73IVGLHH310 pKa = 5.34IAGGTGTEE318 pKa = 3.8NYY320 pKa = 7.1MASVHH325 pKa = 6.56EE326 pKa = 4.05IHH328 pKa = 6.49QLRR331 pKa = 11.84RR332 pKa = 11.84KK333 pKa = 9.93LGLIPEE339 pKa = 4.35VPMVAEE345 pKa = 4.97ASPDD349 pKa = 3.32MKK351 pKa = 10.45HH352 pKa = 6.02RR353 pKa = 11.84QFDD356 pKa = 3.56RR357 pKa = 11.84HH358 pKa = 4.99EE359 pKa = 5.07HH360 pKa = 5.99NDD362 pKa = 3.48LMDD365 pKa = 4.7DD366 pKa = 4.15LSRR369 pKa = 11.84VSKK372 pKa = 10.77HH373 pKa = 5.63GGLLRR378 pKa = 11.84VYY380 pKa = 10.95DD381 pKa = 4.36DD382 pKa = 4.91LAGNDD387 pKa = 3.77YY388 pKa = 11.27AEE390 pKa = 4.66SADD393 pKa = 3.39IPKK396 pKa = 10.64AVGTPVTGLEE406 pKa = 4.4PEE408 pKa = 4.25VSSGSGADD416 pKa = 3.91LVPALAPVPSTGNASAKK433 pKa = 9.66EE434 pKa = 3.97KK435 pKa = 10.49AASEE439 pKa = 4.32VADD442 pKa = 3.61VSTDD446 pKa = 3.13ADD448 pKa = 3.89LSHH451 pKa = 7.44PDD453 pKa = 3.79EE454 pKa = 5.16VPDD457 pKa = 3.96EE458 pKa = 4.44LDD460 pKa = 3.3FSKK463 pKa = 10.82LSEE466 pKa = 4.43EE467 pKa = 4.41EE468 pKa = 4.03PVPPIGVKK476 pKa = 9.82RR477 pKa = 11.84VRR479 pKa = 11.84NKK481 pKa = 10.28KK482 pKa = 9.91VGGIGAFLGTTALAGAVIGKK502 pKa = 9.46VGSAFDD508 pKa = 4.77PKK510 pKa = 10.59WLDD513 pKa = 3.28CTAASLAEE521 pKa = 4.1ASAASYY527 pKa = 10.55HH528 pKa = 7.16AMLNSPNFDD537 pKa = 3.19RR538 pKa = 11.84YY539 pKa = 10.04RR540 pKa = 11.84EE541 pKa = 3.94YY542 pKa = 10.32MSASGVVAADD552 pKa = 3.44QQRR555 pKa = 11.84TMKK558 pKa = 10.75GVDD561 pKa = 3.31GTDD564 pKa = 3.24LARR567 pKa = 11.84LFGTGVSNRR576 pKa = 11.84SRR578 pKa = 11.84KK579 pKa = 9.3KK580 pKa = 9.73ALKK583 pKa = 9.81RR584 pKa = 11.84LPARR588 pKa = 11.84FLEE591 pKa = 4.5VVEE594 pKa = 4.65KK595 pKa = 11.05LGLPVKK601 pKa = 10.48QYY603 pKa = 11.15AQWTLPPAGVEE614 pKa = 4.07AMEE617 pKa = 5.0HH618 pKa = 5.91SLATQLGRR626 pKa = 11.84TCSSCWPEE634 pKa = 3.5EE635 pKa = 3.89TRR637 pKa = 11.84RR638 pKa = 11.84QFANKK643 pKa = 9.93SGPQYY648 pKa = 10.89DD649 pKa = 3.33IFLEE653 pKa = 4.54EE654 pKa = 4.11VARR657 pKa = 11.84YY658 pKa = 6.92PANRR662 pKa = 11.84ADD664 pKa = 3.54AFANVHH670 pKa = 5.71SKK672 pKa = 7.45VTRR675 pKa = 11.84FVLGLDD681 pKa = 3.78GDD683 pKa = 4.54KK684 pKa = 11.1SAGWSQHH691 pKa = 4.84FRR693 pKa = 11.84PGTKK697 pKa = 9.77RR698 pKa = 11.84SWQDD702 pKa = 3.21TEE704 pKa = 4.36GLTLASYY711 pKa = 7.6LTRR714 pKa = 11.84CRR716 pKa = 11.84LLLRR720 pKa = 11.84AAVGPCVMAQMTPSQLIEE738 pKa = 4.86AGLSDD743 pKa = 4.74PRR745 pKa = 11.84TLFIKK750 pKa = 10.67NEE752 pKa = 3.83PHH754 pKa = 6.67SDD756 pKa = 3.3SKK758 pKa = 11.43VADD761 pKa = 3.84GRR763 pKa = 11.84WRR765 pKa = 11.84LIWGASLVDD774 pKa = 3.41VCVASVTCRR783 pKa = 11.84RR784 pKa = 11.84QDD786 pKa = 3.19KK787 pKa = 11.03LDD789 pKa = 3.34IEE791 pKa = 4.79QYY793 pKa = 10.35QGGPVEE799 pKa = 5.43GGHH802 pKa = 5.23QQAAGLGHH810 pKa = 7.13HH811 pKa = 7.27DD812 pKa = 3.56AGISRR817 pKa = 11.84LCVEE821 pKa = 4.85FQRR824 pKa = 11.84LIDD827 pKa = 3.54TGLQVFDD834 pKa = 4.76SDD836 pKa = 3.96ASGWDD841 pKa = 3.09MSVNRR846 pKa = 11.84DD847 pKa = 3.36SLYY850 pKa = 11.05ADD852 pKa = 3.63ALRR855 pKa = 11.84RR856 pKa = 11.84ILLYY860 pKa = 10.19EE861 pKa = 4.1GRR863 pKa = 11.84GKK865 pKa = 10.65AVFEE869 pKa = 3.98RR870 pKa = 11.84LALCEE875 pKa = 3.72AAANSAHH882 pKa = 5.64VVLIGGNLWEE892 pKa = 4.17VLKK895 pKa = 10.86PGITASGILSTTAQNSFIRR914 pKa = 11.84ALLYY918 pKa = 10.2AFVGVRR924 pKa = 11.84HH925 pKa = 5.84SVVAGDD931 pKa = 4.36DD932 pKa = 3.64GAGARR937 pKa = 11.84EE938 pKa = 3.92HH939 pKa = 7.04GYY941 pKa = 10.84DD942 pKa = 3.29HH943 pKa = 6.9VKK945 pKa = 10.75ALAEE949 pKa = 3.99YY950 pKa = 10.22GPEE953 pKa = 3.82EE954 pKa = 4.21KK955 pKa = 10.18QVNLYY960 pKa = 8.82TADD963 pKa = 3.5SGLEE967 pKa = 4.1FTSHH971 pKa = 5.68RR972 pKa = 11.84MRR974 pKa = 11.84RR975 pKa = 11.84GTDD978 pKa = 3.55GTWSAEE984 pKa = 3.57FLNLGKK990 pKa = 10.53ACARR994 pKa = 11.84LAFGDD999 pKa = 4.3TVTADD1004 pKa = 3.41QLAGLMFCVRR1014 pKa = 11.84NNPAEE1019 pKa = 4.33LKK1021 pKa = 9.44TLGDD1025 pKa = 3.56IAEE1028 pKa = 4.18EE1029 pKa = 4.12MGWPIAGVTPAFLPFLDD1046 pKa = 3.85
Molecular weight: 113.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEA9|A0A1L3KEA9_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 9 OX=1922706 PE=4 SV=1
MM1 pKa = 7.71ANGKK5 pKa = 8.49RR6 pKa = 11.84VKK8 pKa = 10.52KK9 pKa = 10.55KK10 pKa = 8.84IVKK13 pKa = 9.97RR14 pKa = 11.84KK15 pKa = 8.59GNGRR19 pKa = 11.84RR20 pKa = 11.84STADD24 pKa = 2.92QVQAQGTGRR33 pKa = 11.84APRR36 pKa = 11.84VPFGNGGGVKK46 pKa = 9.03STIPRR51 pKa = 11.84LPASCWDD58 pKa = 3.69AFSTAHH64 pKa = 6.57APLPRR69 pKa = 11.84AVGPYY74 pKa = 8.48TVVRR78 pKa = 11.84TTRR81 pKa = 11.84LITTNSRR88 pKa = 11.84FALVGSFARR97 pKa = 11.84FGGTAGAADD106 pKa = 4.89GVPLWSNICCATEE119 pKa = 3.59AAGGSIGSTAATTFYY134 pKa = 10.49AIPSPVPDD142 pKa = 4.48TLTGDD147 pKa = 3.36AGGGTLCPSAISVQVMGYY165 pKa = 10.88DD166 pKa = 3.64SLQNAHH172 pKa = 5.78GQIAAAVCPVRR183 pKa = 11.84MDD185 pKa = 3.54LRR187 pKa = 11.84GSTRR191 pKa = 11.84SWDD194 pKa = 3.49GVGAQFLAYY203 pKa = 9.74FRR205 pKa = 11.84PRR207 pKa = 11.84LMSAGKK213 pKa = 7.97VTLRR217 pKa = 11.84GVQMDD222 pKa = 3.71SHH224 pKa = 7.04PLSMADD230 pKa = 3.11VSQFRR235 pKa = 11.84GVDD238 pKa = 3.7EE239 pKa = 4.38VPEE242 pKa = 4.13ALNPLGTTPNSWSLADD258 pKa = 5.09APYY261 pKa = 9.03DD262 pKa = 3.93TNGWAPMVVYY272 pKa = 10.55NPSSANLSLLITFEE286 pKa = 3.53WRR288 pKa = 11.84VRR290 pKa = 11.84FDD292 pKa = 3.04ISNPAVASHH301 pKa = 5.58SHH303 pKa = 6.45HH304 pKa = 6.23GVSSDD309 pKa = 3.68VSWEE313 pKa = 3.82KK314 pKa = 10.85HH315 pKa = 4.05IAAATRR321 pKa = 11.84QLPGVIDD328 pKa = 3.56IVEE331 pKa = 4.34KK332 pKa = 10.52VANTGMAVASMLRR345 pKa = 11.84GG346 pKa = 3.37
MM1 pKa = 7.71ANGKK5 pKa = 8.49RR6 pKa = 11.84VKK8 pKa = 10.52KK9 pKa = 10.55KK10 pKa = 8.84IVKK13 pKa = 9.97RR14 pKa = 11.84KK15 pKa = 8.59GNGRR19 pKa = 11.84RR20 pKa = 11.84STADD24 pKa = 2.92QVQAQGTGRR33 pKa = 11.84APRR36 pKa = 11.84VPFGNGGGVKK46 pKa = 9.03STIPRR51 pKa = 11.84LPASCWDD58 pKa = 3.69AFSTAHH64 pKa = 6.57APLPRR69 pKa = 11.84AVGPYY74 pKa = 8.48TVVRR78 pKa = 11.84TTRR81 pKa = 11.84LITTNSRR88 pKa = 11.84FALVGSFARR97 pKa = 11.84FGGTAGAADD106 pKa = 4.89GVPLWSNICCATEE119 pKa = 3.59AAGGSIGSTAATTFYY134 pKa = 10.49AIPSPVPDD142 pKa = 4.48TLTGDD147 pKa = 3.36AGGGTLCPSAISVQVMGYY165 pKa = 10.88DD166 pKa = 3.64SLQNAHH172 pKa = 5.78GQIAAAVCPVRR183 pKa = 11.84MDD185 pKa = 3.54LRR187 pKa = 11.84GSTRR191 pKa = 11.84SWDD194 pKa = 3.49GVGAQFLAYY203 pKa = 9.74FRR205 pKa = 11.84PRR207 pKa = 11.84LMSAGKK213 pKa = 7.97VTLRR217 pKa = 11.84GVQMDD222 pKa = 3.71SHH224 pKa = 7.04PLSMADD230 pKa = 3.11VSQFRR235 pKa = 11.84GVDD238 pKa = 3.7EE239 pKa = 4.38VPEE242 pKa = 4.13ALNPLGTTPNSWSLADD258 pKa = 5.09APYY261 pKa = 9.03DD262 pKa = 3.93TNGWAPMVVYY272 pKa = 10.55NPSSANLSLLITFEE286 pKa = 3.53WRR288 pKa = 11.84VRR290 pKa = 11.84FDD292 pKa = 3.04ISNPAVASHH301 pKa = 5.58SHH303 pKa = 6.45HH304 pKa = 6.23GVSSDD309 pKa = 3.68VSWEE313 pKa = 3.82KK314 pKa = 10.85HH315 pKa = 4.05IAAATRR321 pKa = 11.84QLPGVIDD328 pKa = 3.56IVEE331 pKa = 4.34KK332 pKa = 10.52VANTGMAVASMLRR345 pKa = 11.84GG346 pKa = 3.37
Molecular weight: 36.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1392 |
346 |
1046 |
696.0 |
74.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.279 ± 0.533 | 1.293 ± 0.071 |
6.034 ± 0.655 | 4.31 ± 1.196 |
3.592 ± 0.192 | 9.986 ± 0.194 |
2.586 ± 0.261 | 3.52 ± 0.11 |
3.951 ± 0.492 | 8.261 ± 1.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.083 ± 0.24 | 2.658 ± 0.51 |
5.029 ± 0.751 | 2.73 ± 0.074 |
6.178 ± 0.218 | 7.471 ± 0.691 |
5.675 ± 0.72 | 8.908 ± 0.024 |
1.509 ± 0.239 | 2.945 ± 0.562 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
