
Nanhai ghost shark arterivirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Nanidovirineae; Nanghoshaviridae; Chimanivirinae; Chimshavirus; Nangarvirus 1
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
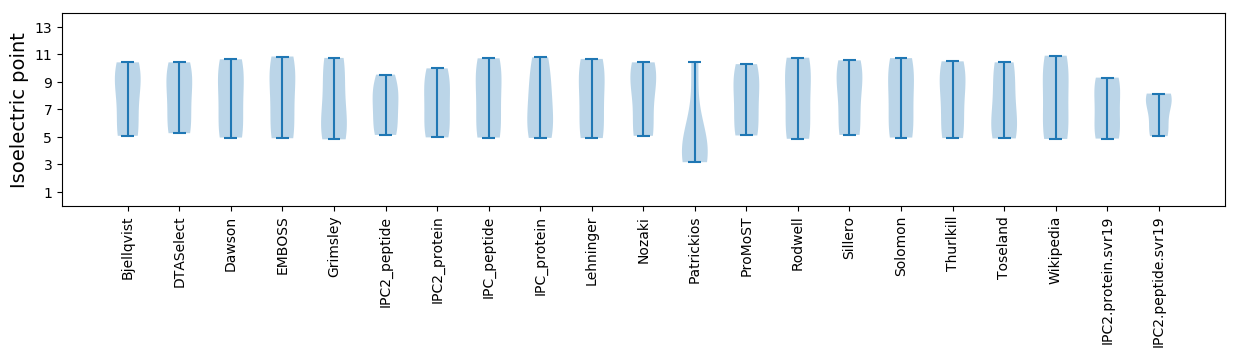
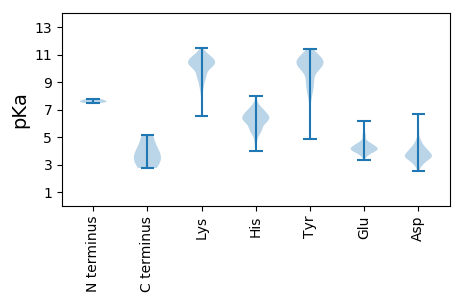
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1GMX9|A0A2P1GMX9_9NIDO Uncharacterized protein OS=Nanhai ghost shark arterivirus OX=2116441 PE=4 SV=1
MM1 pKa = 7.77CCFLLQLVVGCLIYY15 pKa = 10.86LSGVRR20 pKa = 11.84LGVGIPLLEE29 pKa = 4.38SQPLMLTPRR38 pKa = 11.84SFPDD42 pKa = 3.47SSWTAPIQRR51 pKa = 11.84PLTNYY56 pKa = 10.49SDD58 pKa = 4.14LEE60 pKa = 4.53DD61 pKa = 3.59YY62 pKa = 11.02FSFRR66 pKa = 11.84RR67 pKa = 11.84NCTDD71 pKa = 4.85LLDD74 pKa = 4.33HH75 pKa = 6.73MANSHH80 pKa = 5.58MCNEE84 pKa = 4.15TLACLSTLWLGPYY97 pKa = 8.14VTEE100 pKa = 4.5GTCEE104 pKa = 3.77INNLQGASVLAKK116 pKa = 9.97IFRR119 pKa = 11.84RR120 pKa = 11.84VQLAIRR126 pKa = 11.84NEE128 pKa = 4.19NQTPNCTNPGNLMPDD143 pKa = 2.99HH144 pKa = 6.14QTYY147 pKa = 10.19LWLVHH152 pKa = 6.3YY153 pKa = 8.01MPQHH157 pKa = 6.11HH158 pKa = 6.7RR159 pKa = 11.84RR160 pKa = 11.84ADD162 pKa = 3.42LASWLTHH169 pKa = 6.17CRR171 pKa = 11.84ILYY174 pKa = 8.33QRR176 pKa = 11.84EE177 pKa = 4.03YY178 pKa = 8.56THH180 pKa = 7.11PGGFQVQRR188 pKa = 11.84RR189 pKa = 11.84HH190 pKa = 5.7LKK192 pKa = 9.52IFGPFSSTRR201 pKa = 11.84VSGKK205 pKa = 9.32EE206 pKa = 3.65LSGLGLDD213 pKa = 4.72GDD215 pKa = 4.79LFDD218 pKa = 6.69DD219 pKa = 5.33PFDD222 pKa = 5.28GDD224 pKa = 4.94DD225 pKa = 4.09DD226 pKa = 5.76LFDD229 pKa = 5.54FEE231 pKa = 5.17MDD233 pKa = 4.32LDD235 pKa = 4.08PEE237 pKa = 4.26IWSSLVTEE245 pKa = 4.48PPGLEE250 pKa = 4.08LEE252 pKa = 4.67EE253 pKa = 5.36GIQPFPTNLSLTGRR267 pKa = 11.84DD268 pKa = 3.41DD269 pKa = 4.03LEE271 pKa = 3.86IRR273 pKa = 11.84KK274 pKa = 8.78FLDD277 pKa = 3.05NLQRR281 pKa = 11.84PQVYY285 pKa = 8.78TSAWTPKK292 pKa = 9.74HH293 pKa = 6.09PEE295 pKa = 3.53INEE298 pKa = 3.95QAFSAIRR305 pKa = 3.4
MM1 pKa = 7.77CCFLLQLVVGCLIYY15 pKa = 10.86LSGVRR20 pKa = 11.84LGVGIPLLEE29 pKa = 4.38SQPLMLTPRR38 pKa = 11.84SFPDD42 pKa = 3.47SSWTAPIQRR51 pKa = 11.84PLTNYY56 pKa = 10.49SDD58 pKa = 4.14LEE60 pKa = 4.53DD61 pKa = 3.59YY62 pKa = 11.02FSFRR66 pKa = 11.84RR67 pKa = 11.84NCTDD71 pKa = 4.85LLDD74 pKa = 4.33HH75 pKa = 6.73MANSHH80 pKa = 5.58MCNEE84 pKa = 4.15TLACLSTLWLGPYY97 pKa = 8.14VTEE100 pKa = 4.5GTCEE104 pKa = 3.77INNLQGASVLAKK116 pKa = 9.97IFRR119 pKa = 11.84RR120 pKa = 11.84VQLAIRR126 pKa = 11.84NEE128 pKa = 4.19NQTPNCTNPGNLMPDD143 pKa = 2.99HH144 pKa = 6.14QTYY147 pKa = 10.19LWLVHH152 pKa = 6.3YY153 pKa = 8.01MPQHH157 pKa = 6.11HH158 pKa = 6.7RR159 pKa = 11.84RR160 pKa = 11.84ADD162 pKa = 3.42LASWLTHH169 pKa = 6.17CRR171 pKa = 11.84ILYY174 pKa = 8.33QRR176 pKa = 11.84EE177 pKa = 4.03YY178 pKa = 8.56THH180 pKa = 7.11PGGFQVQRR188 pKa = 11.84RR189 pKa = 11.84HH190 pKa = 5.7LKK192 pKa = 9.52IFGPFSSTRR201 pKa = 11.84VSGKK205 pKa = 9.32EE206 pKa = 3.65LSGLGLDD213 pKa = 4.72GDD215 pKa = 4.79LFDD218 pKa = 6.69DD219 pKa = 5.33PFDD222 pKa = 5.28GDD224 pKa = 4.94DD225 pKa = 4.09DD226 pKa = 5.76LFDD229 pKa = 5.54FEE231 pKa = 5.17MDD233 pKa = 4.32LDD235 pKa = 4.08PEE237 pKa = 4.26IWSSLVTEE245 pKa = 4.48PPGLEE250 pKa = 4.08LEE252 pKa = 4.67EE253 pKa = 5.36GIQPFPTNLSLTGRR267 pKa = 11.84DD268 pKa = 3.41DD269 pKa = 4.03LEE271 pKa = 3.86IRR273 pKa = 11.84KK274 pKa = 8.78FLDD277 pKa = 3.05NLQRR281 pKa = 11.84PQVYY285 pKa = 8.78TSAWTPKK292 pKa = 9.74HH293 pKa = 6.09PEE295 pKa = 3.53INEE298 pKa = 3.95QAFSAIRR305 pKa = 3.4
Molecular weight: 34.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1GND3|A0A2P1GND3_9NIDO Uncharacterized protein OS=Nanhai ghost shark arterivirus OX=2116441 PE=4 SV=1
MM1 pKa = 7.57TSFPSWTPEE10 pKa = 3.88ANSPNANNRR19 pKa = 11.84PFPPPPPVQVVYY31 pKa = 11.1VRR33 pKa = 11.84GGGGNWRR40 pKa = 11.84GGRR43 pKa = 11.84GRR45 pKa = 11.84SRR47 pKa = 11.84GRR49 pKa = 11.84GRR51 pKa = 11.84GYY53 pKa = 10.7GYY55 pKa = 10.45NQAPPQQQSPCRR67 pKa = 11.84TIPGFHH73 pKa = 6.62NFNGKK78 pKa = 8.69WYY80 pKa = 8.8PNDD83 pKa = 3.23SGYY86 pKa = 8.93TPKK89 pKa = 10.57QLGEE93 pKa = 4.38SLSNMHH99 pKa = 6.82KK100 pKa = 10.75ASTSEE105 pKa = 4.14HH106 pKa = 5.37NN107 pKa = 3.51
MM1 pKa = 7.57TSFPSWTPEE10 pKa = 3.88ANSPNANNRR19 pKa = 11.84PFPPPPPVQVVYY31 pKa = 11.1VRR33 pKa = 11.84GGGGNWRR40 pKa = 11.84GGRR43 pKa = 11.84GRR45 pKa = 11.84SRR47 pKa = 11.84GRR49 pKa = 11.84GRR51 pKa = 11.84GYY53 pKa = 10.7GYY55 pKa = 10.45NQAPPQQQSPCRR67 pKa = 11.84TIPGFHH73 pKa = 6.62NFNGKK78 pKa = 8.69WYY80 pKa = 8.8PNDD83 pKa = 3.23SGYY86 pKa = 8.93TPKK89 pKa = 10.57QLGEE93 pKa = 4.38SLSNMHH99 pKa = 6.82KK100 pKa = 10.75ASTSEE105 pKa = 4.14HH106 pKa = 5.37NN107 pKa = 3.51
Molecular weight: 11.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4251 |
107 |
3399 |
850.2 |
93.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.187 ± 0.857 | 2.117 ± 0.339 |
3.67 ± 0.617 | 4.846 ± 0.367 |
3.411 ± 0.728 | 7.551 ± 0.642 |
2.376 ± 0.325 | 4.54 ± 0.428 |
4.093 ± 0.639 | 12.021 ± 1.23 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.423 ± 0.174 | 2.893 ± 1.047 |
7.01 ± 0.64 | 3.693 ± 0.353 |
4.869 ± 0.601 | 7.222 ± 0.39 |
7.575 ± 0.46 | 8.516 ± 1.133 |
2.047 ± 0.542 | 2.94 ± 0.388 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
