
Marinithermus hydrothermalis (strain DSM 14884 / JCM 11576 / T1)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Deinococcus-Thermus; Deinococci; Thermales; Thermaceae; Marinithermus; Marinithermus hydrothermalis
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
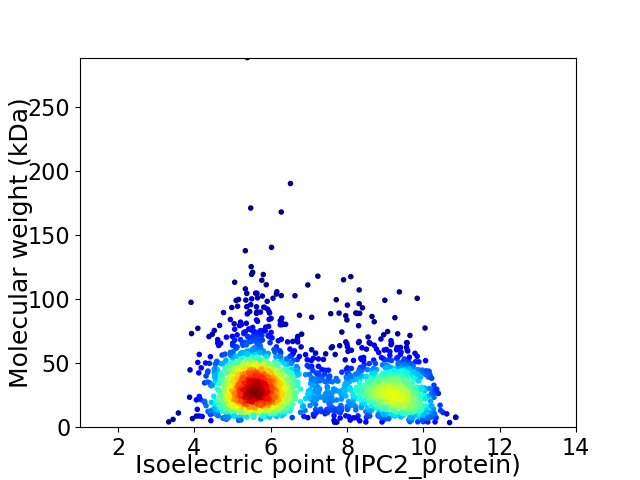
Virtual 2D-PAGE plot for 2194 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2NQ84|F2NQ84_MARHT Transposase IS4 family protein OS=Marinithermus hydrothermalis (strain DSM 14884 / JCM 11576 / T1) OX=869210 GN=Marky_0645 PE=4 SV=1
MM1 pKa = 7.62RR2 pKa = 11.84RR3 pKa = 11.84ILLISVLGLLLAACNQNATSGSKK26 pKa = 9.65PAPSPKK32 pKa = 9.3NAKK35 pKa = 8.3VTVSVTFPEE44 pKa = 4.79SPSEE48 pKa = 3.97APYY51 pKa = 10.85SGLSSIWEE59 pKa = 4.25LFSNDD64 pKa = 3.93PKK66 pKa = 10.97DD67 pKa = 3.69IQPTGAPSSSQKK79 pKa = 11.11AVITVYY85 pKa = 10.53EE86 pKa = 4.45GSTPVTSATVDD97 pKa = 3.33RR98 pKa = 11.84YY99 pKa = 11.29NPTATVLLPAGKK111 pKa = 9.72SYY113 pKa = 10.89RR114 pKa = 11.84FEE116 pKa = 5.63ASVKK120 pKa = 10.18DD121 pKa = 3.88YY122 pKa = 11.58SDD124 pKa = 4.93VEE126 pKa = 4.65VAWGEE131 pKa = 3.86ATQAISGDD139 pKa = 3.64TSVTLSVQTILRR151 pKa = 11.84SALLIPVGGLTANNSEE167 pKa = 4.25VDD169 pKa = 3.62VFIYY173 pKa = 10.17PLTGITGASVPATDD187 pKa = 3.95YY188 pKa = 10.88EE189 pKa = 4.36VTYY192 pKa = 9.07TVQGGTILDD201 pKa = 3.55QNKK204 pKa = 9.84RR205 pKa = 11.84GVRR208 pKa = 11.84IAWDD212 pKa = 3.83GTSDD216 pKa = 3.78PVTVTAQVSGLGEE229 pKa = 4.03DD230 pKa = 4.16HH231 pKa = 7.05NPVTIEE237 pKa = 3.61RR238 pKa = 11.84VYY240 pKa = 11.06NITPATGQGAPLSITGLLPNWPSPFQAKK268 pKa = 7.46VTALDD273 pKa = 3.54VFGAPIYY280 pKa = 6.85EE281 pKa = 4.45TPVGTDD287 pKa = 3.18GSFSLQLPGGDD298 pKa = 3.84ALTPIAIDD306 pKa = 3.64LSEE309 pKa = 4.72LFAVLPQAGCTVFWTTEE326 pKa = 3.71PAPLLNATLVPGQFWIKK343 pKa = 10.77DD344 pKa = 3.56QFTDD348 pKa = 3.34SHH350 pKa = 7.32LGTALLTVSGYY361 pKa = 8.78TLDD364 pKa = 4.02AAFLYY369 pKa = 10.64VDD371 pKa = 4.49RR372 pKa = 11.84SVQAEE377 pKa = 4.16GNATCIAPDD386 pKa = 3.59DD387 pKa = 4.77RR388 pKa = 11.84IITVTVNLNLEE399 pKa = 4.49AGWNLVVSTGDD410 pKa = 3.24PYY412 pKa = 11.65SYY414 pKa = 10.11LTVTGTPLNGSFQAPYY430 pKa = 5.92TTPYY434 pKa = 10.36HH435 pKa = 6.27VIDD438 pKa = 4.29WTLQWHH444 pKa = 4.91TTITDD449 pKa = 3.85LSISAVPQSWAPSEE463 pKa = 4.43GGSFYY468 pKa = 10.82STFTVTNNGPTSLSSVQVALDD489 pKa = 3.69VPSGVTNFYY498 pKa = 11.28VDD500 pKa = 4.13VPNGWFDD507 pKa = 3.49SYY509 pKa = 12.02SNIVYY514 pKa = 10.12LYY516 pKa = 10.84DD517 pKa = 3.69SLAPGDD523 pKa = 3.76NTSFTVNGTTTSGTAGQTLTLSATIQTDD551 pKa = 3.62YY552 pKa = 10.96LTDD555 pKa = 3.55TNPNDD560 pKa = 3.12NTAIVTITPQGGSNANVSVGMDD582 pKa = 4.6LDD584 pKa = 4.52PPAVWFLSPSPGEE597 pKa = 4.33VIPQGSTYY605 pKa = 9.68TVQIEE610 pKa = 4.38VWDD613 pKa = 4.12SSSGEE618 pKa = 4.06SSTPDD623 pKa = 3.01VTEE626 pKa = 3.95IEE628 pKa = 4.92LYY630 pKa = 10.82DD631 pKa = 3.64GAQRR635 pKa = 11.84LGALSSGEE643 pKa = 3.84IVFNPTTNAYY653 pKa = 9.67EE654 pKa = 4.26FVWDD658 pKa = 4.38LTTASPKK665 pKa = 9.9RR666 pKa = 11.84HH667 pKa = 5.76YY668 pKa = 9.93LTVFAYY674 pKa = 7.95DD675 pKa = 3.27TAGNVGQAEE684 pKa = 4.26ISVQVQQ690 pKa = 3.02
MM1 pKa = 7.62RR2 pKa = 11.84RR3 pKa = 11.84ILLISVLGLLLAACNQNATSGSKK26 pKa = 9.65PAPSPKK32 pKa = 9.3NAKK35 pKa = 8.3VTVSVTFPEE44 pKa = 4.79SPSEE48 pKa = 3.97APYY51 pKa = 10.85SGLSSIWEE59 pKa = 4.25LFSNDD64 pKa = 3.93PKK66 pKa = 10.97DD67 pKa = 3.69IQPTGAPSSSQKK79 pKa = 11.11AVITVYY85 pKa = 10.53EE86 pKa = 4.45GSTPVTSATVDD97 pKa = 3.33RR98 pKa = 11.84YY99 pKa = 11.29NPTATVLLPAGKK111 pKa = 9.72SYY113 pKa = 10.89RR114 pKa = 11.84FEE116 pKa = 5.63ASVKK120 pKa = 10.18DD121 pKa = 3.88YY122 pKa = 11.58SDD124 pKa = 4.93VEE126 pKa = 4.65VAWGEE131 pKa = 3.86ATQAISGDD139 pKa = 3.64TSVTLSVQTILRR151 pKa = 11.84SALLIPVGGLTANNSEE167 pKa = 4.25VDD169 pKa = 3.62VFIYY173 pKa = 10.17PLTGITGASVPATDD187 pKa = 3.95YY188 pKa = 10.88EE189 pKa = 4.36VTYY192 pKa = 9.07TVQGGTILDD201 pKa = 3.55QNKK204 pKa = 9.84RR205 pKa = 11.84GVRR208 pKa = 11.84IAWDD212 pKa = 3.83GTSDD216 pKa = 3.78PVTVTAQVSGLGEE229 pKa = 4.03DD230 pKa = 4.16HH231 pKa = 7.05NPVTIEE237 pKa = 3.61RR238 pKa = 11.84VYY240 pKa = 11.06NITPATGQGAPLSITGLLPNWPSPFQAKK268 pKa = 7.46VTALDD273 pKa = 3.54VFGAPIYY280 pKa = 6.85EE281 pKa = 4.45TPVGTDD287 pKa = 3.18GSFSLQLPGGDD298 pKa = 3.84ALTPIAIDD306 pKa = 3.64LSEE309 pKa = 4.72LFAVLPQAGCTVFWTTEE326 pKa = 3.71PAPLLNATLVPGQFWIKK343 pKa = 10.77DD344 pKa = 3.56QFTDD348 pKa = 3.34SHH350 pKa = 7.32LGTALLTVSGYY361 pKa = 8.78TLDD364 pKa = 4.02AAFLYY369 pKa = 10.64VDD371 pKa = 4.49RR372 pKa = 11.84SVQAEE377 pKa = 4.16GNATCIAPDD386 pKa = 3.59DD387 pKa = 4.77RR388 pKa = 11.84IITVTVNLNLEE399 pKa = 4.49AGWNLVVSTGDD410 pKa = 3.24PYY412 pKa = 11.65SYY414 pKa = 10.11LTVTGTPLNGSFQAPYY430 pKa = 5.92TTPYY434 pKa = 10.36HH435 pKa = 6.27VIDD438 pKa = 4.29WTLQWHH444 pKa = 4.91TTITDD449 pKa = 3.85LSISAVPQSWAPSEE463 pKa = 4.43GGSFYY468 pKa = 10.82STFTVTNNGPTSLSSVQVALDD489 pKa = 3.69VPSGVTNFYY498 pKa = 11.28VDD500 pKa = 4.13VPNGWFDD507 pKa = 3.49SYY509 pKa = 12.02SNIVYY514 pKa = 10.12LYY516 pKa = 10.84DD517 pKa = 3.69SLAPGDD523 pKa = 3.76NTSFTVNGTTTSGTAGQTLTLSATIQTDD551 pKa = 3.62YY552 pKa = 10.96LTDD555 pKa = 3.55TNPNDD560 pKa = 3.12NTAIVTITPQGGSNANVSVGMDD582 pKa = 4.6LDD584 pKa = 4.52PPAVWFLSPSPGEE597 pKa = 4.33VIPQGSTYY605 pKa = 9.68TVQIEE610 pKa = 4.38VWDD613 pKa = 4.12SSSGEE618 pKa = 4.06SSTPDD623 pKa = 3.01VTEE626 pKa = 3.95IEE628 pKa = 4.92LYY630 pKa = 10.82DD631 pKa = 3.64GAQRR635 pKa = 11.84LGALSSGEE643 pKa = 3.84IVFNPTTNAYY653 pKa = 9.67EE654 pKa = 4.26FVWDD658 pKa = 4.38LTTASPKK665 pKa = 9.9RR666 pKa = 11.84HH667 pKa = 5.76YY668 pKa = 9.93LTVFAYY674 pKa = 7.95DD675 pKa = 3.27TAGNVGQAEE684 pKa = 4.26ISVQVQQ690 pKa = 3.02
Molecular weight: 73.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2NMZ8|F2NMZ8_MARHT Peptidase M23 OS=Marinithermus hydrothermalis (strain DSM 14884 / JCM 11576 / T1) OX=869210 GN=Marky_2009 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 8.93PTEE5 pKa = 4.94DD6 pKa = 3.87SPSSIVPCPHH16 pKa = 6.68CGATRR21 pKa = 11.84EE22 pKa = 4.07EE23 pKa = 4.22SGGYY27 pKa = 9.02RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 9.3LRR33 pKa = 11.84TFRR36 pKa = 11.84GIQEE40 pKa = 3.99VRR42 pKa = 11.84VKK44 pKa = 10.61RR45 pKa = 11.84IRR47 pKa = 11.84CAQCKK52 pKa = 7.23RR53 pKa = 11.84QKK55 pKa = 10.13RR56 pKa = 11.84ALYY59 pKa = 10.15PEE61 pKa = 4.55DD62 pKa = 3.9CPRR65 pKa = 11.84SRR67 pKa = 11.84WYY69 pKa = 10.42AISIQEE75 pKa = 4.24HH76 pKa = 5.34FLILASHH83 pKa = 7.28RR84 pKa = 11.84APEE87 pKa = 4.41SVQNDD92 pKa = 3.53LAKK95 pKa = 11.0NLGFPLTRR103 pKa = 11.84PTRR106 pKa = 11.84LRR108 pKa = 11.84WLRR111 pKa = 11.84SAGARR116 pKa = 11.84AKK118 pKa = 10.55RR119 pKa = 11.84LLQRR123 pKa = 11.84EE124 pKa = 3.91NRR126 pKa = 11.84LLRR129 pKa = 11.84GRR131 pKa = 11.84VYY133 pKa = 10.08WGSVDD138 pKa = 2.75EE139 pKa = 4.4WAFGRR144 pKa = 11.84GPKK147 pKa = 10.32GYY149 pKa = 10.35GYY151 pKa = 10.84LYY153 pKa = 10.21FEE155 pKa = 5.12AWTGCPLWGDD165 pKa = 3.73LGHH168 pKa = 6.52RR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84YY172 pKa = 9.06EE173 pKa = 3.84RR174 pKa = 11.84VRR176 pKa = 11.84EE177 pKa = 4.01LLWRR181 pKa = 11.84LPPRR185 pKa = 11.84LGVVSDD191 pKa = 4.46GAQEE195 pKa = 3.93VGEE198 pKa = 3.97ALGWLGRR205 pKa = 11.84RR206 pKa = 11.84LLWARR211 pKa = 11.84CAFHH215 pKa = 7.32LMRR218 pKa = 11.84EE219 pKa = 4.15VRR221 pKa = 11.84AKK223 pKa = 10.43VDD225 pKa = 3.74RR226 pKa = 11.84KK227 pKa = 10.6AVGGDD232 pKa = 3.2TGGSEE237 pKa = 4.33GAAGAWAWGEE247 pKa = 4.05GGVLLEE253 pKa = 4.63GASAPVGG260 pKa = 3.49
MM1 pKa = 7.61KK2 pKa = 8.93PTEE5 pKa = 4.94DD6 pKa = 3.87SPSSIVPCPHH16 pKa = 6.68CGATRR21 pKa = 11.84EE22 pKa = 4.07EE23 pKa = 4.22SGGYY27 pKa = 9.02RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 9.3LRR33 pKa = 11.84TFRR36 pKa = 11.84GIQEE40 pKa = 3.99VRR42 pKa = 11.84VKK44 pKa = 10.61RR45 pKa = 11.84IRR47 pKa = 11.84CAQCKK52 pKa = 7.23RR53 pKa = 11.84QKK55 pKa = 10.13RR56 pKa = 11.84ALYY59 pKa = 10.15PEE61 pKa = 4.55DD62 pKa = 3.9CPRR65 pKa = 11.84SRR67 pKa = 11.84WYY69 pKa = 10.42AISIQEE75 pKa = 4.24HH76 pKa = 5.34FLILASHH83 pKa = 7.28RR84 pKa = 11.84APEE87 pKa = 4.41SVQNDD92 pKa = 3.53LAKK95 pKa = 11.0NLGFPLTRR103 pKa = 11.84PTRR106 pKa = 11.84LRR108 pKa = 11.84WLRR111 pKa = 11.84SAGARR116 pKa = 11.84AKK118 pKa = 10.55RR119 pKa = 11.84LLQRR123 pKa = 11.84EE124 pKa = 3.91NRR126 pKa = 11.84LLRR129 pKa = 11.84GRR131 pKa = 11.84VYY133 pKa = 10.08WGSVDD138 pKa = 2.75EE139 pKa = 4.4WAFGRR144 pKa = 11.84GPKK147 pKa = 10.32GYY149 pKa = 10.35GYY151 pKa = 10.84LYY153 pKa = 10.21FEE155 pKa = 5.12AWTGCPLWGDD165 pKa = 3.73LGHH168 pKa = 6.52RR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84YY172 pKa = 9.06EE173 pKa = 3.84RR174 pKa = 11.84VRR176 pKa = 11.84EE177 pKa = 4.01LLWRR181 pKa = 11.84LPPRR185 pKa = 11.84LGVVSDD191 pKa = 4.46GAQEE195 pKa = 3.93VGEE198 pKa = 3.97ALGWLGRR205 pKa = 11.84RR206 pKa = 11.84LLWARR211 pKa = 11.84CAFHH215 pKa = 7.32LMRR218 pKa = 11.84EE219 pKa = 4.15VRR221 pKa = 11.84AKK223 pKa = 10.43VDD225 pKa = 3.74RR226 pKa = 11.84KK227 pKa = 10.6AVGGDD232 pKa = 3.2TGGSEE237 pKa = 4.33GAAGAWAWGEE247 pKa = 4.05GGVLLEE253 pKa = 4.63GASAPVGG260 pKa = 3.49
Molecular weight: 29.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
688454 |
29 |
2681 |
313.8 |
34.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.313 ± 0.072 | 0.481 ± 0.014 |
4.004 ± 0.034 | 7.698 ± 0.064 |
3.51 ± 0.037 | 8.661 ± 0.058 |
2.083 ± 0.027 | 3.655 ± 0.043 |
2.744 ± 0.041 | 12.341 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.656 ± 0.019 | 1.904 ± 0.027 |
6.143 ± 0.045 | 2.939 ± 0.03 |
8.498 ± 0.052 | 3.516 ± 0.028 |
4.817 ± 0.04 | 8.578 ± 0.05 |
1.569 ± 0.027 | 2.889 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
