
Roseburia intestinalis CAG:13
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Roseburia; environmental samples
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
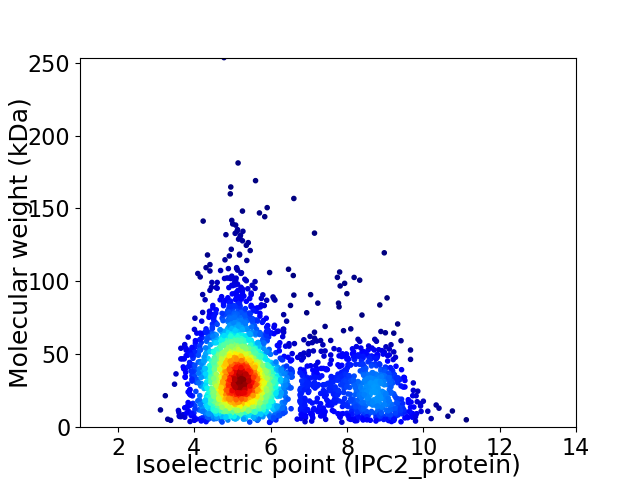
Virtual 2D-PAGE plot for 2856 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
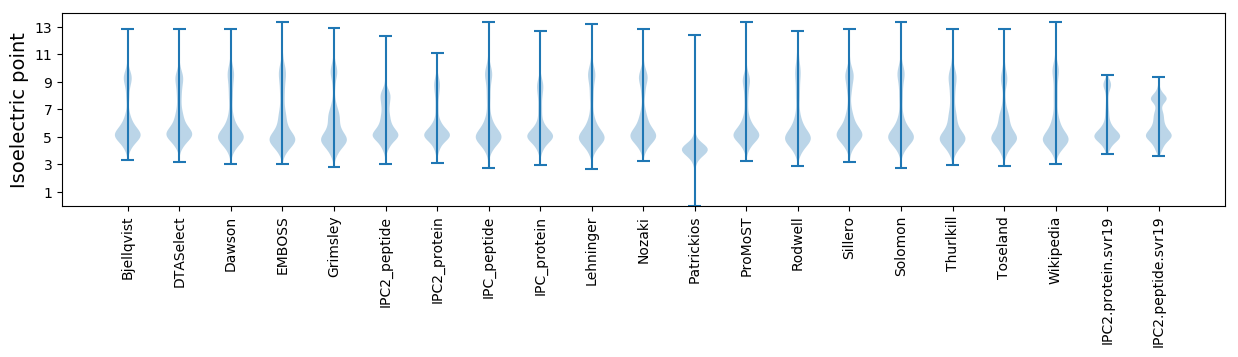
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6AYL2|R6AYL2_9FIRM Uncharacterized protein OS=Roseburia intestinalis CAG:13 OX=1263104 GN=BN484_02606 PE=4 SV=1
MM1 pKa = 7.24KK2 pKa = 10.32KK3 pKa = 10.18KK4 pKa = 10.69VVVLMLCTMMAVTALGCGGKK24 pKa = 10.39KK25 pKa = 10.15NDD27 pKa = 3.74TAEE30 pKa = 4.12GTEE33 pKa = 3.94AAEE36 pKa = 4.36SVNGTEE42 pKa = 4.17TAEE45 pKa = 4.05GTEE48 pKa = 4.24TVEE51 pKa = 4.31YY52 pKa = 10.11EE53 pKa = 4.32SPSYY57 pKa = 11.19DD58 pKa = 4.56LDD60 pKa = 4.16PSDD63 pKa = 4.86YY64 pKa = 10.44VTLCDD69 pKa = 3.73YY70 pKa = 11.43SKK72 pKa = 11.38VPVTITGDD80 pKa = 3.66YY81 pKa = 11.21DD82 pKa = 5.15VDD84 pKa = 3.78DD85 pKa = 4.82QDD87 pKa = 3.85AKK89 pKa = 11.42DD90 pKa = 3.69YY91 pKa = 10.5FEE93 pKa = 5.98QMFSYY98 pKa = 10.73YY99 pKa = 10.96GPFYY103 pKa = 11.03VADD106 pKa = 3.83DD107 pKa = 3.9TKK109 pKa = 9.42TTVGDD114 pKa = 3.66GDD116 pKa = 4.22IVNVDD121 pKa = 3.56YY122 pKa = 11.22VGKK125 pKa = 10.27LDD127 pKa = 5.47GVAFDD132 pKa = 4.43NGSAEE137 pKa = 4.13DD138 pKa = 3.7QNIDD142 pKa = 3.39VDD144 pKa = 4.49NNCSAGSQSSSFIDD158 pKa = 4.73GFTDD162 pKa = 3.53DD163 pKa = 5.28LKK165 pKa = 11.05GASVGDD171 pKa = 4.12VIDD174 pKa = 4.36SDD176 pKa = 4.13VTFPDD181 pKa = 3.2NYY183 pKa = 11.1GNADD187 pKa = 3.7LAGKK191 pKa = 9.3QVVFTFTVNSIQKK204 pKa = 10.05EE205 pKa = 4.15MTLDD209 pKa = 4.28DD210 pKa = 4.13VDD212 pKa = 5.87DD213 pKa = 5.83DD214 pKa = 4.22FAQKK218 pKa = 10.09QFQADD223 pKa = 3.93TVDD226 pKa = 5.17DD227 pKa = 4.25MYY229 pKa = 11.9AQIKK233 pKa = 10.37SFLEE237 pKa = 4.19SQASSNKK244 pKa = 10.38DD245 pKa = 2.81SDD247 pKa = 3.72TYY249 pKa = 10.96TAVQEE254 pKa = 4.18YY255 pKa = 10.58LLEE258 pKa = 4.26NCKK261 pKa = 10.17VDD263 pKa = 3.37IPEE266 pKa = 5.19DD267 pKa = 3.69YY268 pKa = 10.21LTARR272 pKa = 11.84VNDD275 pKa = 3.59YY276 pKa = 10.49EE277 pKa = 4.14KK278 pKa = 10.95QYY280 pKa = 9.39VQNYY284 pKa = 9.16CDD286 pKa = 3.88GDD288 pKa = 3.61ASKK291 pKa = 11.44LEE293 pKa = 4.98DD294 pKa = 3.81YY295 pKa = 11.18VSSQYY300 pKa = 11.91NMTLDD305 pKa = 3.79EE306 pKa = 5.39VRR308 pKa = 11.84QEE310 pKa = 4.01WKK312 pKa = 10.68SGMEE316 pKa = 4.17KK317 pKa = 10.2NISMEE322 pKa = 4.63FITGAIAAKK331 pKa = 10.17EE332 pKa = 4.06GTEE335 pKa = 3.79LDD337 pKa = 3.41EE338 pKa = 6.79DD339 pKa = 4.33GFSSYY344 pKa = 10.62VQTLMSNNSLSSEE357 pKa = 3.81DD358 pKa = 4.87DD359 pKa = 3.43LYY361 pKa = 11.86KK362 pKa = 11.02NYY364 pKa = 10.75GYY366 pKa = 11.26GDD368 pKa = 3.36AAYY371 pKa = 10.06GEE373 pKa = 4.59KK374 pKa = 10.55YY375 pKa = 10.12LRR377 pKa = 11.84QVYY380 pKa = 9.92VDD382 pKa = 4.1NLALQSVKK390 pKa = 11.01DD391 pKa = 3.89NADD394 pKa = 3.4VTVEE398 pKa = 4.23APAEE402 pKa = 4.25TEE404 pKa = 4.08STEE407 pKa = 4.24GTEE410 pKa = 4.11SAEE413 pKa = 3.92PQEE416 pKa = 4.81AVDD419 pKa = 3.58AAEE422 pKa = 4.37TEE424 pKa = 4.46TANN427 pKa = 4.27
MM1 pKa = 7.24KK2 pKa = 10.32KK3 pKa = 10.18KK4 pKa = 10.69VVVLMLCTMMAVTALGCGGKK24 pKa = 10.39KK25 pKa = 10.15NDD27 pKa = 3.74TAEE30 pKa = 4.12GTEE33 pKa = 3.94AAEE36 pKa = 4.36SVNGTEE42 pKa = 4.17TAEE45 pKa = 4.05GTEE48 pKa = 4.24TVEE51 pKa = 4.31YY52 pKa = 10.11EE53 pKa = 4.32SPSYY57 pKa = 11.19DD58 pKa = 4.56LDD60 pKa = 4.16PSDD63 pKa = 4.86YY64 pKa = 10.44VTLCDD69 pKa = 3.73YY70 pKa = 11.43SKK72 pKa = 11.38VPVTITGDD80 pKa = 3.66YY81 pKa = 11.21DD82 pKa = 5.15VDD84 pKa = 3.78DD85 pKa = 4.82QDD87 pKa = 3.85AKK89 pKa = 11.42DD90 pKa = 3.69YY91 pKa = 10.5FEE93 pKa = 5.98QMFSYY98 pKa = 10.73YY99 pKa = 10.96GPFYY103 pKa = 11.03VADD106 pKa = 3.83DD107 pKa = 3.9TKK109 pKa = 9.42TTVGDD114 pKa = 3.66GDD116 pKa = 4.22IVNVDD121 pKa = 3.56YY122 pKa = 11.22VGKK125 pKa = 10.27LDD127 pKa = 5.47GVAFDD132 pKa = 4.43NGSAEE137 pKa = 4.13DD138 pKa = 3.7QNIDD142 pKa = 3.39VDD144 pKa = 4.49NNCSAGSQSSSFIDD158 pKa = 4.73GFTDD162 pKa = 3.53DD163 pKa = 5.28LKK165 pKa = 11.05GASVGDD171 pKa = 4.12VIDD174 pKa = 4.36SDD176 pKa = 4.13VTFPDD181 pKa = 3.2NYY183 pKa = 11.1GNADD187 pKa = 3.7LAGKK191 pKa = 9.3QVVFTFTVNSIQKK204 pKa = 10.05EE205 pKa = 4.15MTLDD209 pKa = 4.28DD210 pKa = 4.13VDD212 pKa = 5.87DD213 pKa = 5.83DD214 pKa = 4.22FAQKK218 pKa = 10.09QFQADD223 pKa = 3.93TVDD226 pKa = 5.17DD227 pKa = 4.25MYY229 pKa = 11.9AQIKK233 pKa = 10.37SFLEE237 pKa = 4.19SQASSNKK244 pKa = 10.38DD245 pKa = 2.81SDD247 pKa = 3.72TYY249 pKa = 10.96TAVQEE254 pKa = 4.18YY255 pKa = 10.58LLEE258 pKa = 4.26NCKK261 pKa = 10.17VDD263 pKa = 3.37IPEE266 pKa = 5.19DD267 pKa = 3.69YY268 pKa = 10.21LTARR272 pKa = 11.84VNDD275 pKa = 3.59YY276 pKa = 10.49EE277 pKa = 4.14KK278 pKa = 10.95QYY280 pKa = 9.39VQNYY284 pKa = 9.16CDD286 pKa = 3.88GDD288 pKa = 3.61ASKK291 pKa = 11.44LEE293 pKa = 4.98DD294 pKa = 3.81YY295 pKa = 11.18VSSQYY300 pKa = 11.91NMTLDD305 pKa = 3.79EE306 pKa = 5.39VRR308 pKa = 11.84QEE310 pKa = 4.01WKK312 pKa = 10.68SGMEE316 pKa = 4.17KK317 pKa = 10.2NISMEE322 pKa = 4.63FITGAIAAKK331 pKa = 10.17EE332 pKa = 4.06GTEE335 pKa = 3.79LDD337 pKa = 3.41EE338 pKa = 6.79DD339 pKa = 4.33GFSSYY344 pKa = 10.62VQTLMSNNSLSSEE357 pKa = 3.81DD358 pKa = 4.87DD359 pKa = 3.43LYY361 pKa = 11.86KK362 pKa = 11.02NYY364 pKa = 10.75GYY366 pKa = 11.26GDD368 pKa = 3.36AAYY371 pKa = 10.06GEE373 pKa = 4.59KK374 pKa = 10.55YY375 pKa = 10.12LRR377 pKa = 11.84QVYY380 pKa = 9.92VDD382 pKa = 4.1NLALQSVKK390 pKa = 11.01DD391 pKa = 3.89NADD394 pKa = 3.4VTVEE398 pKa = 4.23APAEE402 pKa = 4.25TEE404 pKa = 4.08STEE407 pKa = 4.24GTEE410 pKa = 4.11SAEE413 pKa = 3.92PQEE416 pKa = 4.81AVDD419 pKa = 3.58AAEE422 pKa = 4.37TEE424 pKa = 4.46TANN427 pKa = 4.27
Molecular weight: 46.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6B584|R6B584_9FIRM ABC transporter permease/ATP-binding protein OS=Roseburia intestinalis CAG:13 OX=1263104 GN=BN484_02646 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.0VHH16 pKa = 5.93GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.62VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.0VHH16 pKa = 5.93GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.62VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
948991 |
29 |
2374 |
332.3 |
37.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.63 ± 0.049 | 1.476 ± 0.02 |
5.81 ± 0.04 | 7.78 ± 0.053 |
4.08 ± 0.035 | 7.042 ± 0.038 |
1.756 ± 0.019 | 7.257 ± 0.039 |
6.959 ± 0.041 | 8.635 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.231 ± 0.024 | 4.419 ± 0.031 |
3.053 ± 0.022 | 3.272 ± 0.029 |
4.067 ± 0.034 | 5.657 ± 0.037 |
5.612 ± 0.042 | 7.034 ± 0.038 |
0.916 ± 0.016 | 4.308 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
