
Parafilimonas terrae
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Chitinophagia; Chitinophagales; Chitinophagaceae; Parafilimonas
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
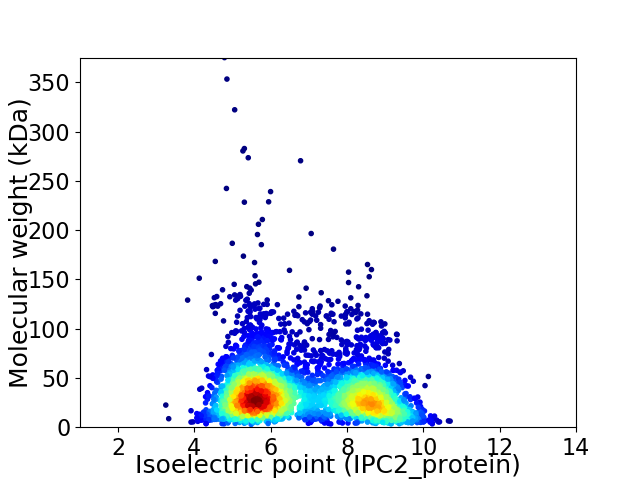
Virtual 2D-PAGE plot for 4249 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I5Y0K7|A0A1I5Y0K7_9BACT Uncharacterized protein OS=Parafilimonas terrae OX=1465490 GN=SAMN05444277_11068 PE=4 SV=1
MM1 pKa = 7.33KK2 pKa = 9.98TRR4 pKa = 11.84LLLFGLSALIVMHH17 pKa = 6.12THH19 pKa = 6.04AQSAGDD25 pKa = 3.8YY26 pKa = 10.08RR27 pKa = 11.84SKK29 pKa = 9.75QTGNWNDD36 pKa = 3.14AANWEE41 pKa = 4.61TFNGTNWVAATNYY54 pKa = 7.13PTATDD59 pKa = 4.15GEE61 pKa = 4.34LSIIATTNITCNVTDD76 pKa = 5.69NIDD79 pKa = 3.39QLTIEE84 pKa = 4.69AGGTLTIDD92 pKa = 3.09NTEE95 pKa = 4.09NFNLTAGPDD104 pKa = 3.56DD105 pKa = 5.37DD106 pKa = 5.34VDD108 pKa = 4.2LKK110 pKa = 11.0IYY112 pKa = 8.0GTLQWNKK119 pKa = 9.23GWAGNCGTWEE129 pKa = 4.1IYY131 pKa = 10.46SGGIANINCGSYY143 pKa = 9.37GTGCNTWNIKK153 pKa = 10.19PGGTMNFIHH162 pKa = 5.57VQCYY166 pKa = 10.45INGTTINNEE175 pKa = 3.55GTINFTPDD183 pKa = 2.86WEE185 pKa = 4.32NMYY188 pKa = 10.32VRR190 pKa = 11.84SNTPNSPGTIHH201 pKa = 6.62NKK203 pKa = 9.83PNGTVNMIKK212 pKa = 9.5TGNNGRR218 pKa = 11.84FGIASDD224 pKa = 4.04GNNGSEE230 pKa = 4.22NYY232 pKa = 10.24VKK234 pKa = 10.61FINEE238 pKa = 3.51GTLNLNEE245 pKa = 4.24GTEE248 pKa = 4.33LYY250 pKa = 10.84YY251 pKa = 10.8GGPGQTSSGISFEE264 pKa = 4.21NTGTINIANNAMLSNAGIFNFNGGNINGTGNALIRR299 pKa = 11.84QIGYY303 pKa = 9.2PGIWKK308 pKa = 10.7SNVPCNLPVNITFEE322 pKa = 4.93LKK324 pKa = 8.89TQLGGPAEE332 pKa = 4.16TTINGKK338 pKa = 9.31LRR340 pKa = 11.84FEE342 pKa = 4.5GRR344 pKa = 11.84LSNRR348 pKa = 11.84DD349 pKa = 3.41YY350 pKa = 11.78SEE352 pKa = 6.11DD353 pKa = 3.36IAKK356 pKa = 10.23LRR358 pKa = 11.84LSPTGTAILVSGYY371 pKa = 10.76LGGAVLTNEE380 pKa = 4.64GVITMPEE387 pKa = 3.61NWGSGLNWEE396 pKa = 5.23GIGKK400 pKa = 9.29FINKK404 pKa = 9.33GLLKK408 pKa = 9.85LTANQTLYY416 pKa = 10.1LHH418 pKa = 6.78NYY420 pKa = 5.33EE421 pKa = 4.55TEE423 pKa = 3.98NGDD426 pKa = 4.09FINEE430 pKa = 4.06GVVEE434 pKa = 4.56IGTGGNLLSNTNSGQTNDD452 pKa = 3.26FFKK455 pKa = 10.37NTSAGIIKK463 pKa = 10.51GNGYY467 pKa = 8.56VQFDD471 pKa = 3.54AGFSNNGAITPGTSPGILQFNGAQPLSANSTLEE504 pKa = 3.87IEE506 pKa = 4.7VKK508 pKa = 10.24DD509 pKa = 3.72ASGAGTGHH517 pKa = 7.0DD518 pKa = 3.3QLTRR522 pKa = 11.84SGDD525 pKa = 3.22LTLAGKK531 pKa = 10.67LEE533 pKa = 4.48VISTGNVPDD542 pKa = 4.18GSYY545 pKa = 10.11MIISLSSGTISGSFSEE561 pKa = 5.39LDD563 pKa = 3.31MPLGYY568 pKa = 8.85TLTVNSDD575 pKa = 3.52NVVLNYY581 pKa = 10.69ASDD584 pKa = 3.62MDD586 pKa = 5.01DD587 pKa = 4.02DD588 pKa = 4.69GVINEE593 pKa = 4.56EE594 pKa = 4.2DD595 pKa = 4.2CAPTDD600 pKa = 3.49ATKK603 pKa = 9.43WQSATLYY610 pKa = 10.2IDD612 pKa = 4.26ADD614 pKa = 3.73GDD616 pKa = 4.13GYY618 pKa = 11.3DD619 pKa = 3.78AGQEE623 pKa = 4.4TVCYY627 pKa = 9.72GATVPDD633 pKa = 5.47GYY635 pKa = 11.23SEE637 pKa = 4.21TSNGEE642 pKa = 3.88DD643 pKa = 5.29CNDD646 pKa = 3.51DD647 pKa = 3.73DD648 pKa = 6.45ANVHH652 pKa = 6.17SSQPYY657 pKa = 8.55YY658 pKa = 11.12ADD660 pKa = 3.32NDD662 pKa = 3.73GDD664 pKa = 4.35GFGDD668 pKa = 3.89AANTTSACSSTPPSGYY684 pKa = 8.51VTNNTDD690 pKa = 5.24CDD692 pKa = 4.46DD693 pKa = 4.39NDD695 pKa = 3.26NTIYY699 pKa = 10.53PGAPEE704 pKa = 5.08LPDD707 pKa = 5.09GKK709 pKa = 10.73DD710 pKa = 3.2NDD712 pKa = 4.51CDD714 pKa = 4.24GEE716 pKa = 4.24TDD718 pKa = 3.51EE719 pKa = 5.74GAGTTWYY726 pKa = 10.35EE727 pKa = 4.18DD728 pKa = 3.46ADD730 pKa = 3.85GDD732 pKa = 4.64GFGNPDD738 pKa = 2.9VSQIAVSQPEE748 pKa = 4.99GYY750 pKa = 10.35VADD753 pKa = 4.33NTDD756 pKa = 3.7CDD758 pKa = 4.14DD759 pKa = 3.91TKK761 pKa = 11.46LLYY764 pKa = 10.58ADD766 pKa = 4.07NDD768 pKa = 3.75GDD770 pKa = 4.2GFGAGSPVACGVDD783 pKa = 3.23NDD785 pKa = 4.73TDD787 pKa = 4.83CDD789 pKa = 4.72DD790 pKa = 4.96NDD792 pKa = 3.68DD793 pKa = 4.41TKK795 pKa = 10.53WQSGLLYY802 pKa = 10.05IDD804 pKa = 4.7FDD806 pKa = 4.82SDD808 pKa = 4.11NFDD811 pKa = 3.47AGQAQVCYY819 pKa = 10.09GAAIPLGYY827 pKa = 10.64NPISLGQDD835 pKa = 3.47CNDD838 pKa = 3.53HH839 pKa = 7.0DD840 pKa = 4.08MHH842 pKa = 7.19VHH844 pKa = 6.12TEE846 pKa = 3.59QTYY849 pKa = 8.77YY850 pKa = 11.37ADD852 pKa = 3.59YY853 pKa = 11.19DD854 pKa = 3.92GDD856 pKa = 4.41GFGDD860 pKa = 3.94PNNSISVCEE869 pKa = 4.47AVPPMYY875 pKa = 10.63YY876 pKa = 8.86VTDD879 pKa = 3.67NTDD882 pKa = 3.89CNDD885 pKa = 3.8NDD887 pKa = 3.36NTIYY891 pKa = 10.52PGAPEE896 pKa = 5.08LPDD899 pKa = 5.09GKK901 pKa = 10.73DD902 pKa = 3.2NDD904 pKa = 4.51CDD906 pKa = 4.25GEE908 pKa = 4.32TDD910 pKa = 3.56EE911 pKa = 5.09GTGTTWYY918 pKa = 10.25EE919 pKa = 4.02DD920 pKa = 3.53ADD922 pKa = 3.85GDD924 pKa = 4.64GFGNPDD930 pKa = 2.9VSQIAVSQPEE940 pKa = 4.99GYY942 pKa = 10.35VADD945 pKa = 4.33NTDD948 pKa = 3.7CDD950 pKa = 4.14DD951 pKa = 3.91TKK953 pKa = 11.46LLYY956 pKa = 10.58ADD958 pKa = 4.07NDD960 pKa = 3.75GDD962 pKa = 4.21GFGAGSLVACGVDD975 pKa = 4.05NNSDD979 pKa = 4.01CNDD982 pKa = 3.56NDD984 pKa = 3.32ATIYY988 pKa = 10.44PGAPEE993 pKa = 4.6LCDD996 pKa = 4.68GKK998 pKa = 10.87DD999 pKa = 3.56NNCDD1003 pKa = 3.01GTTDD1007 pKa = 3.74EE1008 pKa = 5.09GCGITTSLRR1017 pKa = 11.84IDD1019 pKa = 3.68GLAMRR1024 pKa = 11.84EE1025 pKa = 4.28GNSGKK1030 pKa = 10.35RR1031 pKa = 11.84NLNFVITLNKK1041 pKa = 9.41KK1042 pKa = 8.93SNVPITVKK1050 pKa = 10.55YY1051 pKa = 8.23RR1052 pKa = 11.84TTDD1055 pKa = 3.11GSATAPADD1063 pKa = 3.42YY1064 pKa = 10.63VAADD1068 pKa = 3.52KK1069 pKa = 10.84TITIPANTRR1078 pKa = 11.84LRR1080 pKa = 11.84TINIAINGDD1089 pKa = 3.16KK1090 pKa = 10.48DD1091 pKa = 3.65YY1092 pKa = 11.44EE1093 pKa = 4.27PNEE1096 pKa = 4.19TVKK1099 pKa = 10.86VEE1101 pKa = 4.83LYY1103 pKa = 10.6DD1104 pKa = 4.06AVNATIAKK1112 pKa = 7.5STAVGYY1118 pKa = 10.15ILNDD1122 pKa = 3.43DD1123 pKa = 4.31RR1124 pKa = 11.84KK1125 pKa = 9.26QSAVNEE1131 pKa = 4.18EE1132 pKa = 3.97PFAAKK1137 pKa = 9.82AASGGKK1143 pKa = 8.09VTLSPNPASNIITLSGISIGKK1164 pKa = 9.02HH1165 pKa = 3.53ITVSDD1170 pKa = 3.19IKK1172 pKa = 10.99GRR1174 pKa = 11.84TLITQISNASQQKK1187 pKa = 10.2INISNLAAGTYY1198 pKa = 8.16IVRR1201 pKa = 11.84YY1202 pKa = 5.05EE1203 pKa = 4.29TEE1205 pKa = 4.34DD1206 pKa = 3.24GSYY1209 pKa = 11.38NSLKK1213 pKa = 10.51FIKK1216 pKa = 10.19QQ1217 pKa = 3.1
MM1 pKa = 7.33KK2 pKa = 9.98TRR4 pKa = 11.84LLLFGLSALIVMHH17 pKa = 6.12THH19 pKa = 6.04AQSAGDD25 pKa = 3.8YY26 pKa = 10.08RR27 pKa = 11.84SKK29 pKa = 9.75QTGNWNDD36 pKa = 3.14AANWEE41 pKa = 4.61TFNGTNWVAATNYY54 pKa = 7.13PTATDD59 pKa = 4.15GEE61 pKa = 4.34LSIIATTNITCNVTDD76 pKa = 5.69NIDD79 pKa = 3.39QLTIEE84 pKa = 4.69AGGTLTIDD92 pKa = 3.09NTEE95 pKa = 4.09NFNLTAGPDD104 pKa = 3.56DD105 pKa = 5.37DD106 pKa = 5.34VDD108 pKa = 4.2LKK110 pKa = 11.0IYY112 pKa = 8.0GTLQWNKK119 pKa = 9.23GWAGNCGTWEE129 pKa = 4.1IYY131 pKa = 10.46SGGIANINCGSYY143 pKa = 9.37GTGCNTWNIKK153 pKa = 10.19PGGTMNFIHH162 pKa = 5.57VQCYY166 pKa = 10.45INGTTINNEE175 pKa = 3.55GTINFTPDD183 pKa = 2.86WEE185 pKa = 4.32NMYY188 pKa = 10.32VRR190 pKa = 11.84SNTPNSPGTIHH201 pKa = 6.62NKK203 pKa = 9.83PNGTVNMIKK212 pKa = 9.5TGNNGRR218 pKa = 11.84FGIASDD224 pKa = 4.04GNNGSEE230 pKa = 4.22NYY232 pKa = 10.24VKK234 pKa = 10.61FINEE238 pKa = 3.51GTLNLNEE245 pKa = 4.24GTEE248 pKa = 4.33LYY250 pKa = 10.84YY251 pKa = 10.8GGPGQTSSGISFEE264 pKa = 4.21NTGTINIANNAMLSNAGIFNFNGGNINGTGNALIRR299 pKa = 11.84QIGYY303 pKa = 9.2PGIWKK308 pKa = 10.7SNVPCNLPVNITFEE322 pKa = 4.93LKK324 pKa = 8.89TQLGGPAEE332 pKa = 4.16TTINGKK338 pKa = 9.31LRR340 pKa = 11.84FEE342 pKa = 4.5GRR344 pKa = 11.84LSNRR348 pKa = 11.84DD349 pKa = 3.41YY350 pKa = 11.78SEE352 pKa = 6.11DD353 pKa = 3.36IAKK356 pKa = 10.23LRR358 pKa = 11.84LSPTGTAILVSGYY371 pKa = 10.76LGGAVLTNEE380 pKa = 4.64GVITMPEE387 pKa = 3.61NWGSGLNWEE396 pKa = 5.23GIGKK400 pKa = 9.29FINKK404 pKa = 9.33GLLKK408 pKa = 9.85LTANQTLYY416 pKa = 10.1LHH418 pKa = 6.78NYY420 pKa = 5.33EE421 pKa = 4.55TEE423 pKa = 3.98NGDD426 pKa = 4.09FINEE430 pKa = 4.06GVVEE434 pKa = 4.56IGTGGNLLSNTNSGQTNDD452 pKa = 3.26FFKK455 pKa = 10.37NTSAGIIKK463 pKa = 10.51GNGYY467 pKa = 8.56VQFDD471 pKa = 3.54AGFSNNGAITPGTSPGILQFNGAQPLSANSTLEE504 pKa = 3.87IEE506 pKa = 4.7VKK508 pKa = 10.24DD509 pKa = 3.72ASGAGTGHH517 pKa = 7.0DD518 pKa = 3.3QLTRR522 pKa = 11.84SGDD525 pKa = 3.22LTLAGKK531 pKa = 10.67LEE533 pKa = 4.48VISTGNVPDD542 pKa = 4.18GSYY545 pKa = 10.11MIISLSSGTISGSFSEE561 pKa = 5.39LDD563 pKa = 3.31MPLGYY568 pKa = 8.85TLTVNSDD575 pKa = 3.52NVVLNYY581 pKa = 10.69ASDD584 pKa = 3.62MDD586 pKa = 5.01DD587 pKa = 4.02DD588 pKa = 4.69GVINEE593 pKa = 4.56EE594 pKa = 4.2DD595 pKa = 4.2CAPTDD600 pKa = 3.49ATKK603 pKa = 9.43WQSATLYY610 pKa = 10.2IDD612 pKa = 4.26ADD614 pKa = 3.73GDD616 pKa = 4.13GYY618 pKa = 11.3DD619 pKa = 3.78AGQEE623 pKa = 4.4TVCYY627 pKa = 9.72GATVPDD633 pKa = 5.47GYY635 pKa = 11.23SEE637 pKa = 4.21TSNGEE642 pKa = 3.88DD643 pKa = 5.29CNDD646 pKa = 3.51DD647 pKa = 3.73DD648 pKa = 6.45ANVHH652 pKa = 6.17SSQPYY657 pKa = 8.55YY658 pKa = 11.12ADD660 pKa = 3.32NDD662 pKa = 3.73GDD664 pKa = 4.35GFGDD668 pKa = 3.89AANTTSACSSTPPSGYY684 pKa = 8.51VTNNTDD690 pKa = 5.24CDD692 pKa = 4.46DD693 pKa = 4.39NDD695 pKa = 3.26NTIYY699 pKa = 10.53PGAPEE704 pKa = 5.08LPDD707 pKa = 5.09GKK709 pKa = 10.73DD710 pKa = 3.2NDD712 pKa = 4.51CDD714 pKa = 4.24GEE716 pKa = 4.24TDD718 pKa = 3.51EE719 pKa = 5.74GAGTTWYY726 pKa = 10.35EE727 pKa = 4.18DD728 pKa = 3.46ADD730 pKa = 3.85GDD732 pKa = 4.64GFGNPDD738 pKa = 2.9VSQIAVSQPEE748 pKa = 4.99GYY750 pKa = 10.35VADD753 pKa = 4.33NTDD756 pKa = 3.7CDD758 pKa = 4.14DD759 pKa = 3.91TKK761 pKa = 11.46LLYY764 pKa = 10.58ADD766 pKa = 4.07NDD768 pKa = 3.75GDD770 pKa = 4.2GFGAGSPVACGVDD783 pKa = 3.23NDD785 pKa = 4.73TDD787 pKa = 4.83CDD789 pKa = 4.72DD790 pKa = 4.96NDD792 pKa = 3.68DD793 pKa = 4.41TKK795 pKa = 10.53WQSGLLYY802 pKa = 10.05IDD804 pKa = 4.7FDD806 pKa = 4.82SDD808 pKa = 4.11NFDD811 pKa = 3.47AGQAQVCYY819 pKa = 10.09GAAIPLGYY827 pKa = 10.64NPISLGQDD835 pKa = 3.47CNDD838 pKa = 3.53HH839 pKa = 7.0DD840 pKa = 4.08MHH842 pKa = 7.19VHH844 pKa = 6.12TEE846 pKa = 3.59QTYY849 pKa = 8.77YY850 pKa = 11.37ADD852 pKa = 3.59YY853 pKa = 11.19DD854 pKa = 3.92GDD856 pKa = 4.41GFGDD860 pKa = 3.94PNNSISVCEE869 pKa = 4.47AVPPMYY875 pKa = 10.63YY876 pKa = 8.86VTDD879 pKa = 3.67NTDD882 pKa = 3.89CNDD885 pKa = 3.8NDD887 pKa = 3.36NTIYY891 pKa = 10.52PGAPEE896 pKa = 5.08LPDD899 pKa = 5.09GKK901 pKa = 10.73DD902 pKa = 3.2NDD904 pKa = 4.51CDD906 pKa = 4.25GEE908 pKa = 4.32TDD910 pKa = 3.56EE911 pKa = 5.09GTGTTWYY918 pKa = 10.25EE919 pKa = 4.02DD920 pKa = 3.53ADD922 pKa = 3.85GDD924 pKa = 4.64GFGNPDD930 pKa = 2.9VSQIAVSQPEE940 pKa = 4.99GYY942 pKa = 10.35VADD945 pKa = 4.33NTDD948 pKa = 3.7CDD950 pKa = 4.14DD951 pKa = 3.91TKK953 pKa = 11.46LLYY956 pKa = 10.58ADD958 pKa = 4.07NDD960 pKa = 3.75GDD962 pKa = 4.21GFGAGSLVACGVDD975 pKa = 4.05NNSDD979 pKa = 4.01CNDD982 pKa = 3.56NDD984 pKa = 3.32ATIYY988 pKa = 10.44PGAPEE993 pKa = 4.6LCDD996 pKa = 4.68GKK998 pKa = 10.87DD999 pKa = 3.56NNCDD1003 pKa = 3.01GTTDD1007 pKa = 3.74EE1008 pKa = 5.09GCGITTSLRR1017 pKa = 11.84IDD1019 pKa = 3.68GLAMRR1024 pKa = 11.84EE1025 pKa = 4.28GNSGKK1030 pKa = 10.35RR1031 pKa = 11.84NLNFVITLNKK1041 pKa = 9.41KK1042 pKa = 8.93SNVPITVKK1050 pKa = 10.55YY1051 pKa = 8.23RR1052 pKa = 11.84TTDD1055 pKa = 3.11GSATAPADD1063 pKa = 3.42YY1064 pKa = 10.63VAADD1068 pKa = 3.52KK1069 pKa = 10.84TITIPANTRR1078 pKa = 11.84LRR1080 pKa = 11.84TINIAINGDD1089 pKa = 3.16KK1090 pKa = 10.48DD1091 pKa = 3.65YY1092 pKa = 11.44EE1093 pKa = 4.27PNEE1096 pKa = 4.19TVKK1099 pKa = 10.86VEE1101 pKa = 4.83LYY1103 pKa = 10.6DD1104 pKa = 4.06AVNATIAKK1112 pKa = 7.5STAVGYY1118 pKa = 10.15ILNDD1122 pKa = 3.43DD1123 pKa = 4.31RR1124 pKa = 11.84KK1125 pKa = 9.26QSAVNEE1131 pKa = 4.18EE1132 pKa = 3.97PFAAKK1137 pKa = 9.82AASGGKK1143 pKa = 8.09VTLSPNPASNIITLSGISIGKK1164 pKa = 9.02HH1165 pKa = 3.53ITVSDD1170 pKa = 3.19IKK1172 pKa = 10.99GRR1174 pKa = 11.84TLITQISNASQQKK1187 pKa = 10.2INISNLAAGTYY1198 pKa = 8.16IVRR1201 pKa = 11.84YY1202 pKa = 5.05EE1203 pKa = 4.29TEE1205 pKa = 4.34DD1206 pKa = 3.24GSYY1209 pKa = 11.38NSLKK1213 pKa = 10.51FIKK1216 pKa = 10.19QQ1217 pKa = 3.1
Molecular weight: 129.06 kDa
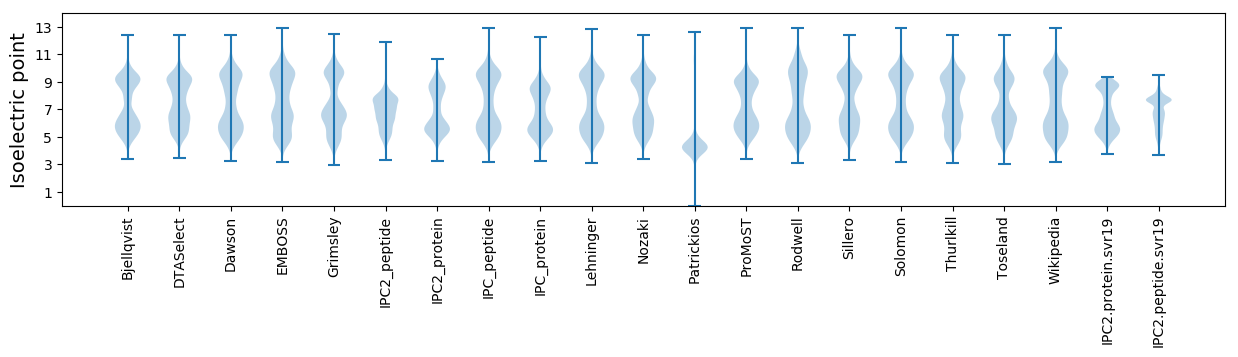
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I5XBR7|A0A1I5XBR7_9BACT N-(5'-phosphoribosyl)anthranilate isomerase OS=Parafilimonas terrae OX=1465490 GN=trpF PE=3 SV=1
MM1 pKa = 7.38KK2 pKa = 9.6RR3 pKa = 11.84TFQPHH8 pKa = 3.73NRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.48SVHH16 pKa = 4.98GFRR19 pKa = 11.84KK20 pKa = 10.11RR21 pKa = 11.84MQTANGRR28 pKa = 11.84KK29 pKa = 8.59VLASRR34 pKa = 11.84RR35 pKa = 11.84TKK37 pKa = 10.38GRR39 pKa = 11.84HH40 pKa = 5.24KK41 pKa = 10.35LTVSDD46 pKa = 3.83EE47 pKa = 4.56RR48 pKa = 11.84KK49 pKa = 10.22LKK51 pKa = 10.73
MM1 pKa = 7.38KK2 pKa = 9.6RR3 pKa = 11.84TFQPHH8 pKa = 3.73NRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.48SVHH16 pKa = 4.98GFRR19 pKa = 11.84KK20 pKa = 10.11RR21 pKa = 11.84MQTANGRR28 pKa = 11.84KK29 pKa = 8.59VLASRR34 pKa = 11.84RR35 pKa = 11.84TKK37 pKa = 10.38GRR39 pKa = 11.84HH40 pKa = 5.24KK41 pKa = 10.35LTVSDD46 pKa = 3.83EE47 pKa = 4.56RR48 pKa = 11.84KK49 pKa = 10.22LKK51 pKa = 10.73
Molecular weight: 6.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1483638 |
25 |
3665 |
349.2 |
39.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.731 ± 0.032 | 0.92 ± 0.013 |
5.431 ± 0.029 | 5.525 ± 0.039 |
4.992 ± 0.028 | 6.578 ± 0.036 |
1.903 ± 0.017 | 7.52 ± 0.036 |
7.244 ± 0.034 | 9.02 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.299 ± 0.019 | 6.22 ± 0.04 |
3.78 ± 0.022 | 3.738 ± 0.023 |
3.592 ± 0.023 | 6.209 ± 0.032 |
5.687 ± 0.047 | 6.053 ± 0.028 |
1.273 ± 0.014 | 4.283 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
