
Papaya cytorhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
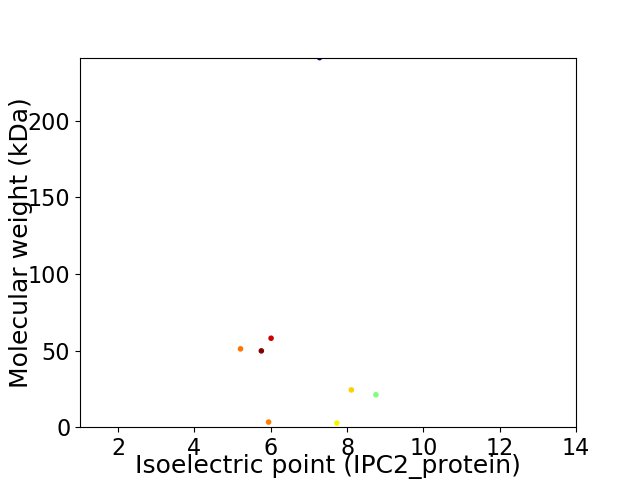
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A386GWS9|A0A386GWS9_9RHAB Large structural protein OS=Papaya cytorhabdovirus OX=2364291 GN=L PE=4 SV=1
MM1 pKa = 7.77PKK3 pKa = 9.59TYY5 pKa = 10.44KK6 pKa = 10.82DD7 pKa = 3.13NSAYY11 pKa = 10.27AKK13 pKa = 10.34EE14 pKa = 4.16IFTPITKK21 pKa = 9.95EE22 pKa = 3.52ISHH25 pKa = 6.35GRR27 pKa = 11.84IQKK30 pKa = 7.46QTWTDD35 pKa = 3.36QVFEE39 pKa = 4.15EE40 pKa = 4.18MQVYY44 pKa = 10.2KK45 pKa = 10.49INKK48 pKa = 6.49YY49 pKa = 7.1TAGDD53 pKa = 3.64LEE55 pKa = 4.72RR56 pKa = 11.84YY57 pKa = 8.46GMYY60 pKa = 9.77MVDD63 pKa = 4.25SISKK67 pKa = 10.2KK68 pKa = 8.57EE69 pKa = 3.9VSMDD73 pKa = 2.84TCYY76 pKa = 10.96AILILACHH84 pKa = 6.24LHH86 pKa = 6.7KK87 pKa = 10.44IAPSKK92 pKa = 11.08DD93 pKa = 3.1FTLFEE98 pKa = 4.84LPLKK102 pKa = 9.79GTLPLPKK109 pKa = 9.34MEE111 pKa = 6.05VFAAWNKK118 pKa = 8.69EE119 pKa = 3.64QAEE122 pKa = 4.32EE123 pKa = 4.45GGEE126 pKa = 3.94EE127 pKa = 4.01DD128 pKa = 3.83GKK130 pKa = 11.31GSEE133 pKa = 4.22EE134 pKa = 4.44SEE136 pKa = 4.02DD137 pKa = 3.78DD138 pKa = 3.98SEE140 pKa = 5.01HH141 pKa = 6.71EE142 pKa = 4.78EE143 pKa = 4.34ISVDD147 pKa = 3.35PSKK150 pKa = 11.15GGKK153 pKa = 9.15EE154 pKa = 3.88KK155 pKa = 10.57TEE157 pKa = 4.51DD158 pKa = 3.8PQSATAIDD166 pKa = 3.76KK167 pKa = 10.31EE168 pKa = 4.19RR169 pKa = 11.84AANCVRR175 pKa = 11.84FFCFCAAFYY184 pKa = 11.06LRR186 pKa = 11.84LCIKK190 pKa = 10.21DD191 pKa = 3.43IQDD194 pKa = 3.23LSTPYY199 pKa = 10.16TRR201 pKa = 11.84MKK203 pKa = 10.18EE204 pKa = 3.85RR205 pKa = 11.84YY206 pKa = 6.74TRR208 pKa = 11.84FYY210 pKa = 10.68QGVIPTSGEE219 pKa = 4.21DD220 pKa = 3.38IPPSEE225 pKa = 4.48SVKK228 pKa = 10.54SLKK231 pKa = 9.89TFFAGDD237 pKa = 4.26RR238 pKa = 11.84IISRR242 pKa = 11.84TWVKK246 pKa = 10.66LFVPWHH252 pKa = 6.91DD253 pKa = 3.57KK254 pKa = 11.71AEE256 pKa = 4.25VGSPQDD262 pKa = 3.09GMVIYY267 pKa = 10.41LAVLPFAFSGMHH279 pKa = 6.44ALKK282 pKa = 11.03LMDD285 pKa = 5.13DD286 pKa = 4.56VIEE289 pKa = 4.17MTHH292 pKa = 6.94LKK294 pKa = 9.39DD295 pKa = 3.08TDD297 pKa = 4.94LIYY300 pKa = 11.16QMMHH304 pKa = 6.54PRR306 pKa = 11.84HH307 pKa = 5.89NSALASISEE316 pKa = 4.77IYY318 pKa = 9.88AKK320 pKa = 10.17HH321 pKa = 5.49YY322 pKa = 10.2PEE324 pKa = 5.43EE325 pKa = 4.86GSQKK329 pKa = 8.74NTYY332 pKa = 8.66VKK334 pKa = 9.43YY335 pKa = 10.57ARR337 pKa = 11.84VVGPQFFPALQTKK350 pKa = 8.68KK351 pKa = 10.68CKK353 pKa = 10.58GLVCVLANLVVLHH366 pKa = 5.96QKK368 pKa = 6.68TTEE371 pKa = 3.93EE372 pKa = 4.09TGDD375 pKa = 3.54PRR377 pKa = 11.84SIAGIDD383 pKa = 4.15DD384 pKa = 3.74ISDD387 pKa = 3.33EE388 pKa = 4.09LMKK391 pKa = 10.63FGKK394 pKa = 10.31RR395 pKa = 11.84MAKK398 pKa = 9.95IMMKK402 pKa = 10.06NAPKK406 pKa = 8.91TASYY410 pKa = 9.27MYY412 pKa = 10.8SSTAKK417 pKa = 9.83EE418 pKa = 3.98AEE420 pKa = 4.22EE421 pKa = 5.29GGDD424 pKa = 3.65STDD427 pKa = 3.66EE428 pKa = 3.88EE429 pKa = 4.78SEE431 pKa = 4.04EE432 pKa = 4.26DD433 pKa = 4.22EE434 pKa = 4.68EE435 pKa = 7.05DD436 pKa = 4.13IDD438 pKa = 5.02PEE440 pKa = 4.14LLKK443 pKa = 10.83VGKK446 pKa = 10.08RR447 pKa = 11.84IYY449 pKa = 10.62KK450 pKa = 9.47PP451 pKa = 3.2
MM1 pKa = 7.77PKK3 pKa = 9.59TYY5 pKa = 10.44KK6 pKa = 10.82DD7 pKa = 3.13NSAYY11 pKa = 10.27AKK13 pKa = 10.34EE14 pKa = 4.16IFTPITKK21 pKa = 9.95EE22 pKa = 3.52ISHH25 pKa = 6.35GRR27 pKa = 11.84IQKK30 pKa = 7.46QTWTDD35 pKa = 3.36QVFEE39 pKa = 4.15EE40 pKa = 4.18MQVYY44 pKa = 10.2KK45 pKa = 10.49INKK48 pKa = 6.49YY49 pKa = 7.1TAGDD53 pKa = 3.64LEE55 pKa = 4.72RR56 pKa = 11.84YY57 pKa = 8.46GMYY60 pKa = 9.77MVDD63 pKa = 4.25SISKK67 pKa = 10.2KK68 pKa = 8.57EE69 pKa = 3.9VSMDD73 pKa = 2.84TCYY76 pKa = 10.96AILILACHH84 pKa = 6.24LHH86 pKa = 6.7KK87 pKa = 10.44IAPSKK92 pKa = 11.08DD93 pKa = 3.1FTLFEE98 pKa = 4.84LPLKK102 pKa = 9.79GTLPLPKK109 pKa = 9.34MEE111 pKa = 6.05VFAAWNKK118 pKa = 8.69EE119 pKa = 3.64QAEE122 pKa = 4.32EE123 pKa = 4.45GGEE126 pKa = 3.94EE127 pKa = 4.01DD128 pKa = 3.83GKK130 pKa = 11.31GSEE133 pKa = 4.22EE134 pKa = 4.44SEE136 pKa = 4.02DD137 pKa = 3.78DD138 pKa = 3.98SEE140 pKa = 5.01HH141 pKa = 6.71EE142 pKa = 4.78EE143 pKa = 4.34ISVDD147 pKa = 3.35PSKK150 pKa = 11.15GGKK153 pKa = 9.15EE154 pKa = 3.88KK155 pKa = 10.57TEE157 pKa = 4.51DD158 pKa = 3.8PQSATAIDD166 pKa = 3.76KK167 pKa = 10.31EE168 pKa = 4.19RR169 pKa = 11.84AANCVRR175 pKa = 11.84FFCFCAAFYY184 pKa = 11.06LRR186 pKa = 11.84LCIKK190 pKa = 10.21DD191 pKa = 3.43IQDD194 pKa = 3.23LSTPYY199 pKa = 10.16TRR201 pKa = 11.84MKK203 pKa = 10.18EE204 pKa = 3.85RR205 pKa = 11.84YY206 pKa = 6.74TRR208 pKa = 11.84FYY210 pKa = 10.68QGVIPTSGEE219 pKa = 4.21DD220 pKa = 3.38IPPSEE225 pKa = 4.48SVKK228 pKa = 10.54SLKK231 pKa = 9.89TFFAGDD237 pKa = 4.26RR238 pKa = 11.84IISRR242 pKa = 11.84TWVKK246 pKa = 10.66LFVPWHH252 pKa = 6.91DD253 pKa = 3.57KK254 pKa = 11.71AEE256 pKa = 4.25VGSPQDD262 pKa = 3.09GMVIYY267 pKa = 10.41LAVLPFAFSGMHH279 pKa = 6.44ALKK282 pKa = 11.03LMDD285 pKa = 5.13DD286 pKa = 4.56VIEE289 pKa = 4.17MTHH292 pKa = 6.94LKK294 pKa = 9.39DD295 pKa = 3.08TDD297 pKa = 4.94LIYY300 pKa = 11.16QMMHH304 pKa = 6.54PRR306 pKa = 11.84HH307 pKa = 5.89NSALASISEE316 pKa = 4.77IYY318 pKa = 9.88AKK320 pKa = 10.17HH321 pKa = 5.49YY322 pKa = 10.2PEE324 pKa = 5.43EE325 pKa = 4.86GSQKK329 pKa = 8.74NTYY332 pKa = 8.66VKK334 pKa = 9.43YY335 pKa = 10.57ARR337 pKa = 11.84VVGPQFFPALQTKK350 pKa = 8.68KK351 pKa = 10.68CKK353 pKa = 10.58GLVCVLANLVVLHH366 pKa = 5.96QKK368 pKa = 6.68TTEE371 pKa = 3.93EE372 pKa = 4.09TGDD375 pKa = 3.54PRR377 pKa = 11.84SIAGIDD383 pKa = 4.15DD384 pKa = 3.74ISDD387 pKa = 3.33EE388 pKa = 4.09LMKK391 pKa = 10.63FGKK394 pKa = 10.31RR395 pKa = 11.84MAKK398 pKa = 9.95IMMKK402 pKa = 10.06NAPKK406 pKa = 8.91TASYY410 pKa = 9.27MYY412 pKa = 10.8SSTAKK417 pKa = 9.83EE418 pKa = 3.98AEE420 pKa = 4.22EE421 pKa = 5.29GGDD424 pKa = 3.65STDD427 pKa = 3.66EE428 pKa = 3.88EE429 pKa = 4.78SEE431 pKa = 4.04EE432 pKa = 4.26DD433 pKa = 4.22EE434 pKa = 4.68EE435 pKa = 7.05DD436 pKa = 4.13IDD438 pKa = 5.02PEE440 pKa = 4.14LLKK443 pKa = 10.83VGKK446 pKa = 10.08RR447 pKa = 11.84IYY449 pKa = 10.62KK450 pKa = 9.47PP451 pKa = 3.2
Molecular weight: 51.15 kDa
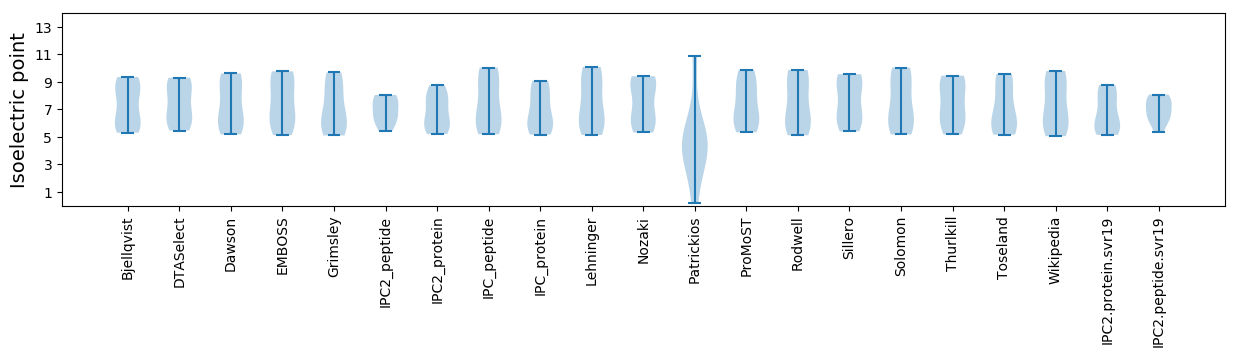
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A386GV82|A0A386GV82_9RHAB Nucleocapsid protein OS=Papaya cytorhabdovirus OX=2364291 GN=N PE=4 SV=1
MM1 pKa = 7.54EE2 pKa = 5.76KK3 pKa = 10.58SNSAKK8 pKa = 10.58GPISSLVSAHH18 pKa = 5.46VSSSKK23 pKa = 10.21RR24 pKa = 11.84IKK26 pKa = 10.95VKK28 pKa = 10.14VQQHH32 pKa = 4.1QEE34 pKa = 4.0EE35 pKa = 4.7YY36 pKa = 10.6SIRR39 pKa = 11.84LPFSTKK45 pKa = 10.47KK46 pKa = 9.55LTSEE50 pKa = 4.32YY51 pKa = 10.47ATVTLLEE58 pKa = 4.29IFYY61 pKa = 10.55KK62 pKa = 10.88GYY64 pKa = 10.31IFNAQGSTIEE74 pKa = 3.87VVIRR78 pKa = 11.84DD79 pKa = 3.96KK80 pKa = 11.31RR81 pKa = 11.84SVSQTEE87 pKa = 3.95QKK89 pKa = 10.17RR90 pKa = 11.84LAVIIPASKK99 pKa = 10.1SCYY102 pKa = 8.49LTVKK106 pKa = 9.82GFSSAPIEE114 pKa = 4.19EE115 pKa = 4.61GCPYY119 pKa = 9.92EE120 pKa = 4.89VSIKK124 pKa = 9.82TVVRR128 pKa = 11.84NIRR131 pKa = 11.84FGITLGRR138 pKa = 11.84LDD140 pKa = 3.7IRR142 pKa = 11.84PHH144 pKa = 6.27LFYY147 pKa = 11.11SNHH150 pKa = 5.94FEE152 pKa = 4.68DD153 pKa = 5.47GDD155 pKa = 4.0YY156 pKa = 10.72LTVNKK161 pKa = 10.04VEE163 pKa = 4.2GPEE166 pKa = 4.08LSLKK170 pKa = 10.58RR171 pKa = 11.84GSLTNKK177 pKa = 7.13EE178 pKa = 3.94APYY181 pKa = 9.34TVSLIPTEE189 pKa = 4.02
MM1 pKa = 7.54EE2 pKa = 5.76KK3 pKa = 10.58SNSAKK8 pKa = 10.58GPISSLVSAHH18 pKa = 5.46VSSSKK23 pKa = 10.21RR24 pKa = 11.84IKK26 pKa = 10.95VKK28 pKa = 10.14VQQHH32 pKa = 4.1QEE34 pKa = 4.0EE35 pKa = 4.7YY36 pKa = 10.6SIRR39 pKa = 11.84LPFSTKK45 pKa = 10.47KK46 pKa = 9.55LTSEE50 pKa = 4.32YY51 pKa = 10.47ATVTLLEE58 pKa = 4.29IFYY61 pKa = 10.55KK62 pKa = 10.88GYY64 pKa = 10.31IFNAQGSTIEE74 pKa = 3.87VVIRR78 pKa = 11.84DD79 pKa = 3.96KK80 pKa = 11.31RR81 pKa = 11.84SVSQTEE87 pKa = 3.95QKK89 pKa = 10.17RR90 pKa = 11.84LAVIIPASKK99 pKa = 10.1SCYY102 pKa = 8.49LTVKK106 pKa = 9.82GFSSAPIEE114 pKa = 4.19EE115 pKa = 4.61GCPYY119 pKa = 9.92EE120 pKa = 4.89VSIKK124 pKa = 9.82TVVRR128 pKa = 11.84NIRR131 pKa = 11.84FGITLGRR138 pKa = 11.84LDD140 pKa = 3.7IRR142 pKa = 11.84PHH144 pKa = 6.27LFYY147 pKa = 11.11SNHH150 pKa = 5.94FEE152 pKa = 4.68DD153 pKa = 5.47GDD155 pKa = 4.0YY156 pKa = 10.72LTVNKK161 pKa = 10.04VEE163 pKa = 4.2GPEE166 pKa = 4.08LSLKK170 pKa = 10.58RR171 pKa = 11.84GSLTNKK177 pKa = 7.13EE178 pKa = 3.94APYY181 pKa = 9.34TVSLIPTEE189 pKa = 4.02
Molecular weight: 21.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3985 |
26 |
2113 |
498.1 |
56.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.868 ± 0.557 | 1.807 ± 0.379 |
5.445 ± 0.527 | 7.202 ± 1.142 |
4.768 ± 0.7 | 5.32 ± 0.186 |
1.907 ± 0.18 | 6.349 ± 0.22 |
7.955 ± 1.183 | 9.26 ± 0.879 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.061 ± 0.297 | 3.614 ± 0.4 |
4.567 ± 0.258 | 2.886 ± 0.321 |
4.592 ± 0.463 | 9.184 ± 0.55 |
5.621 ± 0.244 | 6.851 ± 0.394 |
1.305 ± 0.29 | 3.438 ± 0.282 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
