
Southern cowpea mosaic virus (SCPMV) (Southern bean mosaic virus (strain cowpea))
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Sobemovirus
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
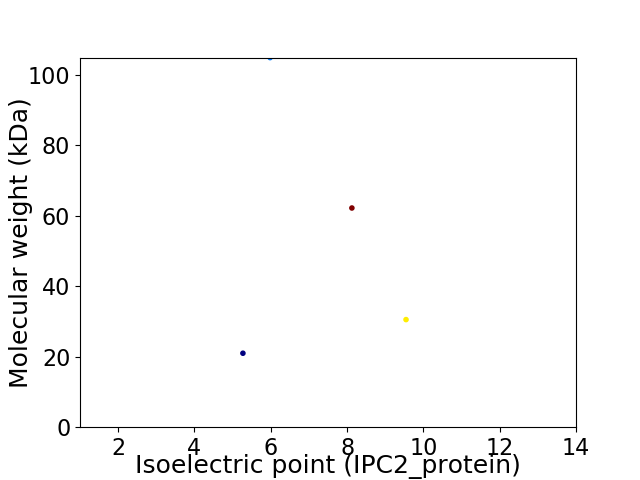
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
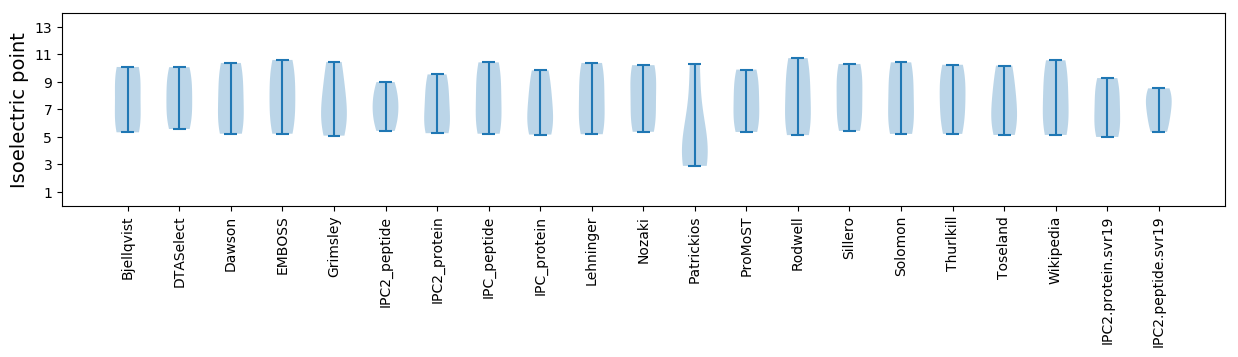
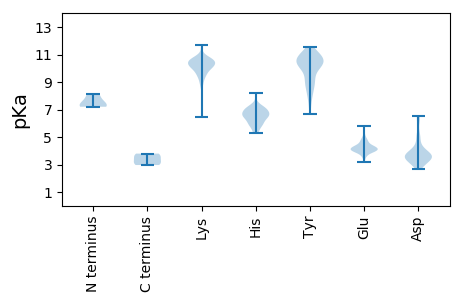
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q83470|P2A_SCPMV Polyprotein P2A OS=Southern cowpea mosaic virus OX=196398 GN=ORF2A PE=4 SV=2
MM1 pKa = 8.13IYY3 pKa = 8.18EE4 pKa = 4.29TLVLSKK10 pKa = 10.13TEE12 pKa = 4.04LEE14 pKa = 4.21QLNVDD19 pKa = 4.31LPRR22 pKa = 11.84YY23 pKa = 9.16SYY25 pKa = 10.79RR26 pKa = 11.84FRR28 pKa = 11.84YY29 pKa = 9.09SRR31 pKa = 11.84SIGDD35 pKa = 3.66TVVEE39 pKa = 4.65FPGTLSDD46 pKa = 3.42PCIPVVDD53 pKa = 4.28VLLGACWAWPQPSRR67 pKa = 11.84HH68 pKa = 6.08GGLGLDD74 pKa = 5.42DD75 pKa = 5.15IDD77 pKa = 4.94PFDD80 pKa = 5.06ASFSCCVTSPEE91 pKa = 4.47RR92 pKa = 11.84YY93 pKa = 8.55CLSRR97 pKa = 11.84SVLSGVDD104 pKa = 3.22AFVVRR109 pKa = 11.84GSCKK113 pKa = 10.06LCGLGFLDD121 pKa = 3.99NFNPFEE127 pKa = 4.03IRR129 pKa = 11.84ALLGQTTPGLWWQPVKK145 pKa = 10.34PVYY148 pKa = 10.21DD149 pKa = 4.02DD150 pKa = 4.79RR151 pKa = 11.84NIHH154 pKa = 6.46LPYY157 pKa = 10.51DD158 pKa = 3.58SEE160 pKa = 4.49LNARR164 pKa = 11.84IQRR167 pKa = 11.84FQYY170 pKa = 8.84TCRR173 pKa = 11.84EE174 pKa = 4.28CIVRR178 pKa = 11.84VAFHH182 pKa = 6.23MSSS185 pKa = 2.96
MM1 pKa = 8.13IYY3 pKa = 8.18EE4 pKa = 4.29TLVLSKK10 pKa = 10.13TEE12 pKa = 4.04LEE14 pKa = 4.21QLNVDD19 pKa = 4.31LPRR22 pKa = 11.84YY23 pKa = 9.16SYY25 pKa = 10.79RR26 pKa = 11.84FRR28 pKa = 11.84YY29 pKa = 9.09SRR31 pKa = 11.84SIGDD35 pKa = 3.66TVVEE39 pKa = 4.65FPGTLSDD46 pKa = 3.42PCIPVVDD53 pKa = 4.28VLLGACWAWPQPSRR67 pKa = 11.84HH68 pKa = 6.08GGLGLDD74 pKa = 5.42DD75 pKa = 5.15IDD77 pKa = 4.94PFDD80 pKa = 5.06ASFSCCVTSPEE91 pKa = 4.47RR92 pKa = 11.84YY93 pKa = 8.55CLSRR97 pKa = 11.84SVLSGVDD104 pKa = 3.22AFVVRR109 pKa = 11.84GSCKK113 pKa = 10.06LCGLGFLDD121 pKa = 3.99NFNPFEE127 pKa = 4.03IRR129 pKa = 11.84ALLGQTTPGLWWQPVKK145 pKa = 10.34PVYY148 pKa = 10.21DD149 pKa = 4.02DD150 pKa = 4.79RR151 pKa = 11.84NIHH154 pKa = 6.46LPYY157 pKa = 10.51DD158 pKa = 3.58SEE160 pKa = 4.49LNARR164 pKa = 11.84IQRR167 pKa = 11.84FQYY170 pKa = 8.84TCRR173 pKa = 11.84EE174 pKa = 4.28CIVRR178 pKa = 11.84VAFHH182 pKa = 6.23MSSS185 pKa = 2.96
Molecular weight: 20.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P21405|RDRP_SCPMV Replicase polyprotein P2AB OS=Southern cowpea mosaic virus OX=196398 GN=ORF2A-2B PE=3 SV=2
MM1 pKa = 7.55SGLFHH6 pKa = 7.07HH7 pKa = 6.26RR8 pKa = 11.84TKK10 pKa = 10.49PRR12 pKa = 11.84EE13 pKa = 3.39IRR15 pKa = 11.84AFVMATRR22 pKa = 11.84LTKK25 pKa = 10.36KK26 pKa = 10.17QLAQAIQNTLPNPPRR41 pKa = 11.84RR42 pKa = 11.84KK43 pKa = 9.39RR44 pKa = 11.84RR45 pKa = 11.84AKK47 pKa = 9.92RR48 pKa = 11.84RR49 pKa = 11.84AAQVPKK55 pKa = 8.55PTQAGVSMAPIAQGTMVKK73 pKa = 10.22LRR75 pKa = 11.84PPMLRR80 pKa = 11.84SSMDD84 pKa = 3.13VTILSHH90 pKa = 7.0CEE92 pKa = 3.6LSTEE96 pKa = 4.11LAVTVTIVVTSEE108 pKa = 3.58LVMPFTVGTWLRR120 pKa = 11.84GVAQNWSKK128 pKa = 10.44YY129 pKa = 7.72AWVAIRR135 pKa = 11.84YY136 pKa = 6.64TYY138 pKa = 10.79LPSCPTTTSGAIHH151 pKa = 6.6MGFQYY156 pKa = 11.58DD157 pKa = 3.61MADD160 pKa = 3.6TLPVSVNQLSNLKK173 pKa = 10.37GYY175 pKa = 7.99VTGPVWEE182 pKa = 4.78GQSGLCFVNNTKK194 pKa = 10.71CPDD197 pKa = 3.1TSRR200 pKa = 11.84AITIALDD207 pKa = 3.54TNEE210 pKa = 4.22VSEE213 pKa = 4.22KK214 pKa = 10.3RR215 pKa = 11.84YY216 pKa = 9.24PFKK219 pKa = 10.48TATDD223 pKa = 3.61YY224 pKa = 10.01ATAVGVNANIGNILVPARR242 pKa = 11.84LVTAMEE248 pKa = 4.81GGSSKK253 pKa = 9.71TAVNTGRR260 pKa = 11.84LYY262 pKa = 10.88ASYY265 pKa = 9.38TIRR268 pKa = 11.84LIEE271 pKa = 4.83PIAAALNLL279 pKa = 3.8
MM1 pKa = 7.55SGLFHH6 pKa = 7.07HH7 pKa = 6.26RR8 pKa = 11.84TKK10 pKa = 10.49PRR12 pKa = 11.84EE13 pKa = 3.39IRR15 pKa = 11.84AFVMATRR22 pKa = 11.84LTKK25 pKa = 10.36KK26 pKa = 10.17QLAQAIQNTLPNPPRR41 pKa = 11.84RR42 pKa = 11.84KK43 pKa = 9.39RR44 pKa = 11.84RR45 pKa = 11.84AKK47 pKa = 9.92RR48 pKa = 11.84RR49 pKa = 11.84AAQVPKK55 pKa = 8.55PTQAGVSMAPIAQGTMVKK73 pKa = 10.22LRR75 pKa = 11.84PPMLRR80 pKa = 11.84SSMDD84 pKa = 3.13VTILSHH90 pKa = 7.0CEE92 pKa = 3.6LSTEE96 pKa = 4.11LAVTVTIVVTSEE108 pKa = 3.58LVMPFTVGTWLRR120 pKa = 11.84GVAQNWSKK128 pKa = 10.44YY129 pKa = 7.72AWVAIRR135 pKa = 11.84YY136 pKa = 6.64TYY138 pKa = 10.79LPSCPTTTSGAIHH151 pKa = 6.6MGFQYY156 pKa = 11.58DD157 pKa = 3.61MADD160 pKa = 3.6TLPVSVNQLSNLKK173 pKa = 10.37GYY175 pKa = 7.99VTGPVWEE182 pKa = 4.78GQSGLCFVNNTKK194 pKa = 10.71CPDD197 pKa = 3.1TSRR200 pKa = 11.84AITIALDD207 pKa = 3.54TNEE210 pKa = 4.22VSEE213 pKa = 4.22KK214 pKa = 10.3RR215 pKa = 11.84YY216 pKa = 9.24PFKK219 pKa = 10.48TATDD223 pKa = 3.61YY224 pKa = 10.01ATAVGVNANIGNILVPARR242 pKa = 11.84LVTAMEE248 pKa = 4.81GGSSKK253 pKa = 9.71TAVNTGRR260 pKa = 11.84LYY262 pKa = 10.88ASYY265 pKa = 9.38TIRR268 pKa = 11.84LIEE271 pKa = 4.83PIAAALNLL279 pKa = 3.8
Molecular weight: 30.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1992 |
185 |
956 |
498.0 |
54.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.082 ± 0.762 | 2.41 ± 0.418 |
4.267 ± 0.474 | 5.221 ± 0.706 |
3.363 ± 0.393 | 7.279 ± 0.471 |
2.058 ± 0.216 | 4.016 ± 0.331 |
4.97 ± 0.662 | 9.036 ± 0.306 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.56 ± 0.289 | 3.313 ± 0.342 |
6.978 ± 0.63 | 3.213 ± 0.096 |
5.321 ± 0.656 | 8.735 ± 0.592 |
6.074 ± 1.078 | 8.133 ± 0.376 |
2.008 ± 0.156 | 2.962 ± 0.293 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
