
Halothiobacillus sp. XI15
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Halothiobacillaceae; Halothiobacillus; unclassified Halothiobacillus
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
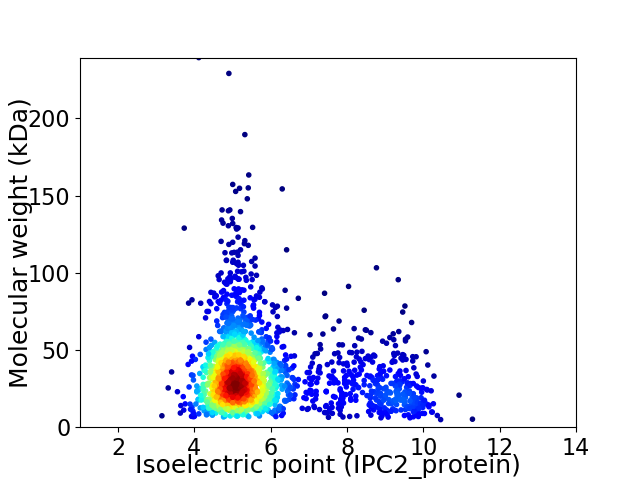
Virtual 2D-PAGE plot for 2012 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0W1RTT8|A0A0W1RTT8_9GAMM HTH arsR-type domain-containing protein OS=Halothiobacillus sp. XI15 OX=1766620 GN=AUR63_02155 PE=4 SV=1
MM1 pKa = 7.62WAGQATAITIDD12 pKa = 3.65QKK14 pKa = 11.37PLIVQQALPPNVMLLLDD31 pKa = 4.8DD32 pKa = 4.4SGSMLRR38 pKa = 11.84EE39 pKa = 3.91YY40 pKa = 10.39MPDD43 pKa = 3.06ASYY46 pKa = 11.48LSDD49 pKa = 3.36TSDD52 pKa = 3.18QGMQSASVNYY62 pKa = 9.36IYY64 pKa = 10.98YY65 pKa = 9.93DD66 pKa = 3.37GSVTYY71 pKa = 9.1EE72 pKa = 4.16PPVKK76 pKa = 10.56ADD78 pKa = 3.03GTYY81 pKa = 10.84YY82 pKa = 11.03SDD84 pKa = 2.82ITTFPTLPLDD94 pKa = 4.22GFDD97 pKa = 4.46SSSSTADD104 pKa = 3.0VTRR107 pKa = 11.84YY108 pKa = 9.79YY109 pKa = 11.57SNGSYY114 pKa = 11.39ANVNFPFFRR123 pKa = 11.84AFCPPDD129 pKa = 3.35TTQSVVHH136 pKa = 6.29YY137 pKa = 8.95GDD139 pKa = 5.0CYY141 pKa = 10.81SATYY145 pKa = 10.71YY146 pKa = 11.27NDD148 pKa = 3.46ADD150 pKa = 4.0HH151 pKa = 6.93VFLFQDD157 pKa = 3.41SGNNLYY163 pKa = 10.71YY164 pKa = 10.83YY165 pKa = 10.23EE166 pKa = 5.66ADD168 pKa = 3.13ALAFVYY174 pKa = 7.51TTPDD178 pKa = 2.87GSGGYY183 pKa = 9.14IEE185 pKa = 5.89NYY187 pKa = 9.73VATNCADD194 pKa = 3.92LPTDD198 pKa = 3.94AQATCADD205 pKa = 4.27DD206 pKa = 5.38AATQQNVGNWFGYY219 pKa = 9.6YY220 pKa = 7.37RR221 pKa = 11.84TRR223 pKa = 11.84MLAAKK228 pKa = 9.48SGLMNGFADD237 pKa = 3.86VSAEE241 pKa = 3.61FRR243 pKa = 11.84VGFGSINGNSADD255 pKa = 3.96YY256 pKa = 10.51IRR258 pKa = 11.84NITAGEE264 pKa = 4.22CGDD267 pKa = 3.95VFSASCRR274 pKa = 11.84YY275 pKa = 10.45GFVTDD280 pKa = 3.62TNNNNEE286 pKa = 4.29LAGVATWGDD295 pKa = 3.52GAAGTRR301 pKa = 11.84KK302 pKa = 10.35DD303 pKa = 4.52EE304 pKa = 4.27FWGWLEE310 pKa = 5.33DD311 pKa = 4.55AGGDD315 pKa = 3.41QNTPLRR321 pKa = 11.84EE322 pKa = 3.94TLEE325 pKa = 4.34AAGEE329 pKa = 4.31YY330 pKa = 10.54YY331 pKa = 9.15RR332 pKa = 11.84TGQPWSSRR340 pKa = 11.84DD341 pKa = 3.41SSGTSQEE348 pKa = 4.28YY349 pKa = 10.63ACRR352 pKa = 11.84QSYY355 pKa = 8.66TILTSDD361 pKa = 3.77GFWNEE366 pKa = 3.65VGAPDD371 pKa = 4.82PDD373 pKa = 3.56VGNADD378 pKa = 3.76GTSGATLGDD387 pKa = 4.39DD388 pKa = 4.35GSGTMTAPNGIEE400 pKa = 3.89YY401 pKa = 8.75TYY403 pKa = 10.82EE404 pKa = 3.95AVDD407 pKa = 4.37PYY409 pKa = 11.56SDD411 pKa = 4.75DD412 pKa = 3.99FSNTLADD419 pKa = 3.12VAMYY423 pKa = 9.66YY424 pKa = 9.45WKK426 pKa = 10.59NDD428 pKa = 2.85LRR430 pKa = 11.84TGLTNQVPTTNEE442 pKa = 3.97DD443 pKa = 3.5PAFWQHH449 pKa = 5.0MTTFAIGLGYY459 pKa = 10.64DD460 pKa = 3.56PLDD463 pKa = 3.97ANGNVIDD470 pKa = 4.47SDD472 pKa = 5.32AVFDD476 pKa = 4.04WANGGDD482 pKa = 4.34PISGFAWPEE491 pKa = 3.73PASGSINNIADD502 pKa = 3.7MLHH505 pKa = 6.37AAVNGHH511 pKa = 6.08GGFYY515 pKa = 10.12SAKK518 pKa = 9.87SSQAFAAAISDD529 pKa = 3.76AVGRR533 pKa = 11.84TAEE536 pKa = 4.16RR537 pKa = 11.84VGTGASLAANSTRR550 pKa = 11.84LEE552 pKa = 4.4TGTVTYY558 pKa = 9.42QANYY562 pKa = 7.51FTGTWVGDD570 pKa = 3.61LKK572 pKa = 11.03AYY574 pKa = 9.68PVDD577 pKa = 3.98PLTKK581 pKa = 10.34QIATYY586 pKa = 9.32PDD588 pKa = 3.2WSAADD593 pKa = 3.63NLLAEE598 pKa = 4.93PADD601 pKa = 3.74SCAASAGEE609 pKa = 3.97VCPDD613 pKa = 2.49GRR615 pKa = 11.84AIYY618 pKa = 9.0TYY620 pKa = 9.74ATSDD624 pKa = 3.45ANLGNNDD631 pKa = 3.13PQYY634 pKa = 11.22RR635 pKa = 11.84EE636 pKa = 4.01FLASDD641 pKa = 3.63VASLSNAQQAALGGTTTEE659 pKa = 3.92RR660 pKa = 11.84QAIVDD665 pKa = 4.18FLRR668 pKa = 11.84GDD670 pKa = 3.62TTNAEE675 pKa = 4.44ANGGNYY681 pKa = 9.0RR682 pKa = 11.84DD683 pKa = 3.92RR684 pKa = 11.84STALGDD690 pKa = 3.53IVNSQPVYY698 pKa = 10.58VGQPDD703 pKa = 4.38PNLHH707 pKa = 5.5TARR710 pKa = 11.84SFDD713 pKa = 3.31GVEE716 pKa = 4.76FYY718 pKa = 10.43PAFAEE723 pKa = 5.2DD724 pKa = 4.37PEE726 pKa = 5.86GDD728 pKa = 4.05NDD730 pKa = 3.77NQGIDD735 pKa = 3.84PEE737 pKa = 4.93GGSNTRR743 pKa = 11.84EE744 pKa = 3.82PLVYY748 pKa = 10.24VAANDD753 pKa = 3.87GMLHH757 pKa = 6.27GFDD760 pKa = 5.48AITGEE765 pKa = 4.1EE766 pKa = 5.15LYY768 pKa = 11.1AFMPGAVIEE777 pKa = 4.53AGVKK781 pKa = 10.08KK782 pKa = 10.43LADD785 pKa = 4.02PDD787 pKa = 3.74YY788 pKa = 10.7GANPGHH794 pKa = 6.41EE795 pKa = 4.16FFNDD799 pKa = 3.76GEE801 pKa = 4.43LTVADD806 pKa = 4.38VYY808 pKa = 11.27IDD810 pKa = 3.88LPYY813 pKa = 10.94TNGSDD818 pKa = 3.91VQWRR822 pKa = 11.84TVLVGTTGRR831 pKa = 11.84GEE833 pKa = 4.28AKK835 pKa = 9.27TVYY838 pKa = 10.55ALDD841 pKa = 3.35ITDD844 pKa = 4.05PADD847 pKa = 4.47IRR849 pKa = 11.84FLWEE853 pKa = 3.7RR854 pKa = 11.84SAGDD858 pKa = 3.25GGANAGYY865 pKa = 9.15IGQMIGKK872 pKa = 8.34PLVVQTANGEE882 pKa = 4.05WSVLIGNGYY891 pKa = 10.46NSANDD896 pKa = 3.57SAALLQFDD904 pKa = 4.53VGTGALDD911 pKa = 3.31VHH913 pKa = 5.58VTNDD917 pKa = 4.0GITGNGLAAPATFDD931 pKa = 3.74YY932 pKa = 10.62TDD934 pKa = 4.22SPDD937 pKa = 4.2GIQDD941 pKa = 3.2VAYY944 pKa = 10.42AGDD947 pKa = 3.78LNGWVWRR954 pKa = 11.84FDD956 pKa = 3.73LSTSTSNGVQVYY968 pKa = 8.84QATRR972 pKa = 11.84ADD974 pKa = 3.75GTAQPITAGLLLGKK988 pKa = 10.19NPATSNLWAFFGTGQYY1004 pKa = 10.8LSQGDD1009 pKa = 4.08LTDD1012 pKa = 3.63QSVQTWYY1019 pKa = 11.25GLIVEE1024 pKa = 4.87SADD1027 pKa = 3.08TSLPVSSGDD1036 pKa = 3.33GRR1038 pKa = 11.84SVLVEE1043 pKa = 3.67RR1044 pKa = 11.84DD1045 pKa = 3.33IVDD1048 pKa = 3.57EE1049 pKa = 4.0QSNGTFTARR1058 pKa = 11.84AFSQGAANDD1067 pKa = 3.74MDD1069 pKa = 5.18GKK1071 pKa = 10.49SGWYY1075 pKa = 9.04IDD1077 pKa = 5.91LISPDD1082 pKa = 3.36NGAEE1086 pKa = 4.2GEE1088 pKa = 4.4RR1089 pKa = 11.84MVEE1092 pKa = 3.84PNQFQGEE1099 pKa = 4.59VLVGTSVIPDD1109 pKa = 3.42AADD1112 pKa = 2.84ICNPSGRR1119 pKa = 11.84GFIMGIDD1126 pKa = 3.82PFDD1129 pKa = 3.83GTNVEE1134 pKa = 4.29QAFVDD1139 pKa = 3.89YY1140 pKa = 11.29NGDD1143 pKa = 3.78GVVDD1147 pKa = 4.64GSDD1150 pKa = 3.98VITGDD1155 pKa = 4.05DD1156 pKa = 3.6GVTYY1160 pKa = 10.63AVGAIGFNRR1169 pKa = 11.84LPNAPIFVGNTMLISFDD1186 pKa = 4.27DD1187 pKa = 4.52GSTASVEE1194 pKa = 4.14AAGGGSSGQRR1204 pKa = 11.84SSWRR1208 pKa = 11.84EE1209 pKa = 3.57IIFDD1213 pKa = 3.63
MM1 pKa = 7.62WAGQATAITIDD12 pKa = 3.65QKK14 pKa = 11.37PLIVQQALPPNVMLLLDD31 pKa = 4.8DD32 pKa = 4.4SGSMLRR38 pKa = 11.84EE39 pKa = 3.91YY40 pKa = 10.39MPDD43 pKa = 3.06ASYY46 pKa = 11.48LSDD49 pKa = 3.36TSDD52 pKa = 3.18QGMQSASVNYY62 pKa = 9.36IYY64 pKa = 10.98YY65 pKa = 9.93DD66 pKa = 3.37GSVTYY71 pKa = 9.1EE72 pKa = 4.16PPVKK76 pKa = 10.56ADD78 pKa = 3.03GTYY81 pKa = 10.84YY82 pKa = 11.03SDD84 pKa = 2.82ITTFPTLPLDD94 pKa = 4.22GFDD97 pKa = 4.46SSSSTADD104 pKa = 3.0VTRR107 pKa = 11.84YY108 pKa = 9.79YY109 pKa = 11.57SNGSYY114 pKa = 11.39ANVNFPFFRR123 pKa = 11.84AFCPPDD129 pKa = 3.35TTQSVVHH136 pKa = 6.29YY137 pKa = 8.95GDD139 pKa = 5.0CYY141 pKa = 10.81SATYY145 pKa = 10.71YY146 pKa = 11.27NDD148 pKa = 3.46ADD150 pKa = 4.0HH151 pKa = 6.93VFLFQDD157 pKa = 3.41SGNNLYY163 pKa = 10.71YY164 pKa = 10.83YY165 pKa = 10.23EE166 pKa = 5.66ADD168 pKa = 3.13ALAFVYY174 pKa = 7.51TTPDD178 pKa = 2.87GSGGYY183 pKa = 9.14IEE185 pKa = 5.89NYY187 pKa = 9.73VATNCADD194 pKa = 3.92LPTDD198 pKa = 3.94AQATCADD205 pKa = 4.27DD206 pKa = 5.38AATQQNVGNWFGYY219 pKa = 9.6YY220 pKa = 7.37RR221 pKa = 11.84TRR223 pKa = 11.84MLAAKK228 pKa = 9.48SGLMNGFADD237 pKa = 3.86VSAEE241 pKa = 3.61FRR243 pKa = 11.84VGFGSINGNSADD255 pKa = 3.96YY256 pKa = 10.51IRR258 pKa = 11.84NITAGEE264 pKa = 4.22CGDD267 pKa = 3.95VFSASCRR274 pKa = 11.84YY275 pKa = 10.45GFVTDD280 pKa = 3.62TNNNNEE286 pKa = 4.29LAGVATWGDD295 pKa = 3.52GAAGTRR301 pKa = 11.84KK302 pKa = 10.35DD303 pKa = 4.52EE304 pKa = 4.27FWGWLEE310 pKa = 5.33DD311 pKa = 4.55AGGDD315 pKa = 3.41QNTPLRR321 pKa = 11.84EE322 pKa = 3.94TLEE325 pKa = 4.34AAGEE329 pKa = 4.31YY330 pKa = 10.54YY331 pKa = 9.15RR332 pKa = 11.84TGQPWSSRR340 pKa = 11.84DD341 pKa = 3.41SSGTSQEE348 pKa = 4.28YY349 pKa = 10.63ACRR352 pKa = 11.84QSYY355 pKa = 8.66TILTSDD361 pKa = 3.77GFWNEE366 pKa = 3.65VGAPDD371 pKa = 4.82PDD373 pKa = 3.56VGNADD378 pKa = 3.76GTSGATLGDD387 pKa = 4.39DD388 pKa = 4.35GSGTMTAPNGIEE400 pKa = 3.89YY401 pKa = 8.75TYY403 pKa = 10.82EE404 pKa = 3.95AVDD407 pKa = 4.37PYY409 pKa = 11.56SDD411 pKa = 4.75DD412 pKa = 3.99FSNTLADD419 pKa = 3.12VAMYY423 pKa = 9.66YY424 pKa = 9.45WKK426 pKa = 10.59NDD428 pKa = 2.85LRR430 pKa = 11.84TGLTNQVPTTNEE442 pKa = 3.97DD443 pKa = 3.5PAFWQHH449 pKa = 5.0MTTFAIGLGYY459 pKa = 10.64DD460 pKa = 3.56PLDD463 pKa = 3.97ANGNVIDD470 pKa = 4.47SDD472 pKa = 5.32AVFDD476 pKa = 4.04WANGGDD482 pKa = 4.34PISGFAWPEE491 pKa = 3.73PASGSINNIADD502 pKa = 3.7MLHH505 pKa = 6.37AAVNGHH511 pKa = 6.08GGFYY515 pKa = 10.12SAKK518 pKa = 9.87SSQAFAAAISDD529 pKa = 3.76AVGRR533 pKa = 11.84TAEE536 pKa = 4.16RR537 pKa = 11.84VGTGASLAANSTRR550 pKa = 11.84LEE552 pKa = 4.4TGTVTYY558 pKa = 9.42QANYY562 pKa = 7.51FTGTWVGDD570 pKa = 3.61LKK572 pKa = 11.03AYY574 pKa = 9.68PVDD577 pKa = 3.98PLTKK581 pKa = 10.34QIATYY586 pKa = 9.32PDD588 pKa = 3.2WSAADD593 pKa = 3.63NLLAEE598 pKa = 4.93PADD601 pKa = 3.74SCAASAGEE609 pKa = 3.97VCPDD613 pKa = 2.49GRR615 pKa = 11.84AIYY618 pKa = 9.0TYY620 pKa = 9.74ATSDD624 pKa = 3.45ANLGNNDD631 pKa = 3.13PQYY634 pKa = 11.22RR635 pKa = 11.84EE636 pKa = 4.01FLASDD641 pKa = 3.63VASLSNAQQAALGGTTTEE659 pKa = 3.92RR660 pKa = 11.84QAIVDD665 pKa = 4.18FLRR668 pKa = 11.84GDD670 pKa = 3.62TTNAEE675 pKa = 4.44ANGGNYY681 pKa = 9.0RR682 pKa = 11.84DD683 pKa = 3.92RR684 pKa = 11.84STALGDD690 pKa = 3.53IVNSQPVYY698 pKa = 10.58VGQPDD703 pKa = 4.38PNLHH707 pKa = 5.5TARR710 pKa = 11.84SFDD713 pKa = 3.31GVEE716 pKa = 4.76FYY718 pKa = 10.43PAFAEE723 pKa = 5.2DD724 pKa = 4.37PEE726 pKa = 5.86GDD728 pKa = 4.05NDD730 pKa = 3.77NQGIDD735 pKa = 3.84PEE737 pKa = 4.93GGSNTRR743 pKa = 11.84EE744 pKa = 3.82PLVYY748 pKa = 10.24VAANDD753 pKa = 3.87GMLHH757 pKa = 6.27GFDD760 pKa = 5.48AITGEE765 pKa = 4.1EE766 pKa = 5.15LYY768 pKa = 11.1AFMPGAVIEE777 pKa = 4.53AGVKK781 pKa = 10.08KK782 pKa = 10.43LADD785 pKa = 4.02PDD787 pKa = 3.74YY788 pKa = 10.7GANPGHH794 pKa = 6.41EE795 pKa = 4.16FFNDD799 pKa = 3.76GEE801 pKa = 4.43LTVADD806 pKa = 4.38VYY808 pKa = 11.27IDD810 pKa = 3.88LPYY813 pKa = 10.94TNGSDD818 pKa = 3.91VQWRR822 pKa = 11.84TVLVGTTGRR831 pKa = 11.84GEE833 pKa = 4.28AKK835 pKa = 9.27TVYY838 pKa = 10.55ALDD841 pKa = 3.35ITDD844 pKa = 4.05PADD847 pKa = 4.47IRR849 pKa = 11.84FLWEE853 pKa = 3.7RR854 pKa = 11.84SAGDD858 pKa = 3.25GGANAGYY865 pKa = 9.15IGQMIGKK872 pKa = 8.34PLVVQTANGEE882 pKa = 4.05WSVLIGNGYY891 pKa = 10.46NSANDD896 pKa = 3.57SAALLQFDD904 pKa = 4.53VGTGALDD911 pKa = 3.31VHH913 pKa = 5.58VTNDD917 pKa = 4.0GITGNGLAAPATFDD931 pKa = 3.74YY932 pKa = 10.62TDD934 pKa = 4.22SPDD937 pKa = 4.2GIQDD941 pKa = 3.2VAYY944 pKa = 10.42AGDD947 pKa = 3.78LNGWVWRR954 pKa = 11.84FDD956 pKa = 3.73LSTSTSNGVQVYY968 pKa = 8.84QATRR972 pKa = 11.84ADD974 pKa = 3.75GTAQPITAGLLLGKK988 pKa = 10.19NPATSNLWAFFGTGQYY1004 pKa = 10.8LSQGDD1009 pKa = 4.08LTDD1012 pKa = 3.63QSVQTWYY1019 pKa = 11.25GLIVEE1024 pKa = 4.87SADD1027 pKa = 3.08TSLPVSSGDD1036 pKa = 3.33GRR1038 pKa = 11.84SVLVEE1043 pKa = 3.67RR1044 pKa = 11.84DD1045 pKa = 3.33IVDD1048 pKa = 3.57EE1049 pKa = 4.0QSNGTFTARR1058 pKa = 11.84AFSQGAANDD1067 pKa = 3.74MDD1069 pKa = 5.18GKK1071 pKa = 10.49SGWYY1075 pKa = 9.04IDD1077 pKa = 5.91LISPDD1082 pKa = 3.36NGAEE1086 pKa = 4.2GEE1088 pKa = 4.4RR1089 pKa = 11.84MVEE1092 pKa = 3.84PNQFQGEE1099 pKa = 4.59VLVGTSVIPDD1109 pKa = 3.42AADD1112 pKa = 2.84ICNPSGRR1119 pKa = 11.84GFIMGIDD1126 pKa = 3.82PFDD1129 pKa = 3.83GTNVEE1134 pKa = 4.29QAFVDD1139 pKa = 3.89YY1140 pKa = 11.29NGDD1143 pKa = 3.78GVVDD1147 pKa = 4.64GSDD1150 pKa = 3.98VITGDD1155 pKa = 4.05DD1156 pKa = 3.6GVTYY1160 pKa = 10.63AVGAIGFNRR1169 pKa = 11.84LPNAPIFVGNTMLISFDD1186 pKa = 4.27DD1187 pKa = 4.52GSTASVEE1194 pKa = 4.14AAGGGSSGQRR1204 pKa = 11.84SSWRR1208 pKa = 11.84EE1209 pKa = 3.57IIFDD1213 pKa = 3.63
Molecular weight: 128.97 kDa
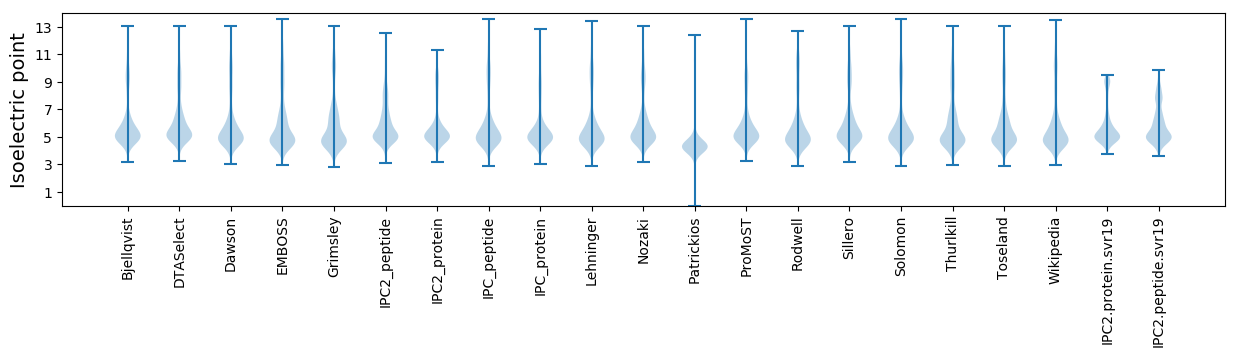
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0W1RSW3|A0A0W1RSW3_9GAMM DNA deoxyribophosphodiesterase OS=Halothiobacillus sp. XI15 OX=1766620 GN=AUR63_03360 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.16RR14 pKa = 11.84THH16 pKa = 6.07GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 9.32TRR25 pKa = 11.84GGRR28 pKa = 11.84AIINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.16RR14 pKa = 11.84THH16 pKa = 6.07GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 9.32TRR25 pKa = 11.84GGRR28 pKa = 11.84AIINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
676788 |
41 |
2201 |
336.4 |
36.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.436 ± 0.073 | 0.711 ± 0.016 |
6.417 ± 0.051 | 6.962 ± 0.055 |
3.449 ± 0.034 | 8.333 ± 0.054 |
2.312 ± 0.027 | 4.894 ± 0.04 |
2.613 ± 0.045 | 10.525 ± 0.075 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.404 ± 0.025 | 2.505 ± 0.029 |
5.049 ± 0.038 | 3.519 ± 0.031 |
7.809 ± 0.054 | 5.033 ± 0.035 |
4.97 ± 0.038 | 7.424 ± 0.056 |
1.426 ± 0.023 | 2.21 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
