
Hubei permutotetra-like virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
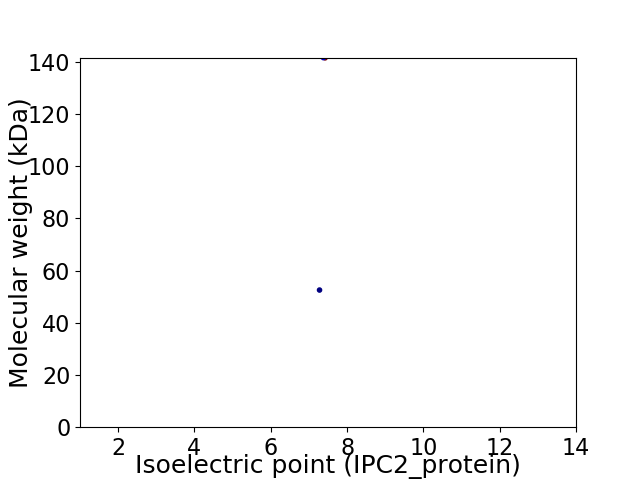
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KHW2|A0A1L3KHW2_9VIRU Putative capsid protein OS=Hubei permutotetra-like virus 6 OX=1923080 PE=4 SV=1
MM1 pKa = 7.59DD2 pKa = 4.07TSNPVVSSDD11 pKa = 2.99RR12 pKa = 11.84RR13 pKa = 11.84SLRR16 pKa = 11.84EE17 pKa = 3.85AKK19 pKa = 10.01EE20 pKa = 3.46LAVVKK25 pKa = 10.31KK26 pKa = 10.43RR27 pKa = 11.84QDD29 pKa = 2.95LEE31 pKa = 4.4YY32 pKa = 10.58IMNVAQTSTRR42 pKa = 11.84VILPVGEE49 pKa = 4.21RR50 pKa = 11.84ALPEE54 pKa = 4.34AEE56 pKa = 4.2VKK58 pKa = 10.74SIILNLKK65 pKa = 10.09DD66 pKa = 3.51KK67 pKa = 10.35EE68 pKa = 4.41PTVRR72 pKa = 11.84TDD74 pKa = 3.33SEE76 pKa = 4.49DD77 pKa = 3.52MLDD80 pKa = 3.33EE81 pKa = 4.44TVFQLVAPSMPVCGLLSQDD100 pKa = 4.22GVPLHH105 pKa = 6.73PPPIHH110 pKa = 5.75NAKK113 pKa = 10.25GRR115 pKa = 11.84VTTNRR120 pKa = 11.84RR121 pKa = 11.84FGGGAYY127 pKa = 9.14KK128 pKa = 10.51PLGIRR133 pKa = 11.84PMVEE137 pKa = 3.1ILGVRR142 pKa = 11.84TQEE145 pKa = 3.42ITKK148 pKa = 9.99VYY150 pKa = 8.91THH152 pKa = 6.38GTWQGFSNRR161 pKa = 11.84LAKK164 pKa = 10.46QMGRR168 pKa = 11.84KK169 pKa = 8.06CVSMGMTIRR178 pKa = 11.84NSMPGVQMDD187 pKa = 3.58KK188 pKa = 9.47NTPRR192 pKa = 11.84RR193 pKa = 11.84IFFDD197 pKa = 3.06ILNKK201 pKa = 10.16KK202 pKa = 8.63MPARR206 pKa = 11.84KK207 pKa = 8.83PSGWPSLSDD216 pKa = 3.79PLDD219 pKa = 4.13TILDD223 pKa = 4.16NIKK226 pKa = 8.76ITLDD230 pKa = 3.26ASAGPPYY237 pKa = 9.5WRR239 pKa = 11.84NKK241 pKa = 8.22VEE243 pKa = 5.39CMDD246 pKa = 4.08QILSEE251 pKa = 4.66GIPLLVKK258 pKa = 10.25ALKK261 pKa = 10.19EE262 pKa = 3.87NRR264 pKa = 11.84LEE266 pKa = 3.87QFMRR270 pKa = 11.84EE271 pKa = 3.8EE272 pKa = 4.59PEE274 pKa = 4.24FFLCEE279 pKa = 4.11VKK281 pKa = 10.95NKK283 pKa = 9.89LDD285 pKa = 3.5RR286 pKa = 11.84YY287 pKa = 10.37GVDD290 pKa = 3.51EE291 pKa = 5.75LDD293 pKa = 3.92VKK295 pKa = 10.58CRR297 pKa = 11.84PYY299 pKa = 9.94ICYY302 pKa = 8.37PAHH305 pKa = 6.61CALLFSVLCQRR316 pKa = 11.84FQEE319 pKa = 4.19KK320 pKa = 11.07LEE322 pKa = 4.1IFTQNFEE329 pKa = 4.01SSNAYY334 pKa = 9.93GFSSANGGLKK344 pKa = 10.37KK345 pKa = 10.35LVEE348 pKa = 4.33WMISTPRR355 pKa = 11.84GTSRR359 pKa = 11.84YY360 pKa = 8.91ICYY363 pKa = 10.56GDD365 pKa = 3.49DD366 pKa = 3.32TRR368 pKa = 11.84IVVADD373 pKa = 4.19RR374 pKa = 11.84NGDD377 pKa = 3.3RR378 pKa = 11.84FMVNPDD384 pKa = 3.72FQQMDD389 pKa = 3.96GSVDD393 pKa = 3.29AQTIAWTIDD402 pKa = 3.5FIIKK406 pKa = 10.27KK407 pKa = 10.32LEE409 pKa = 4.15EE410 pKa = 3.97EE411 pKa = 4.63DD412 pKa = 5.01GEE414 pKa = 4.69EE415 pKa = 3.92NCFWRR420 pKa = 11.84AVAKK424 pKa = 9.94LWKK427 pKa = 10.52LMATNPQFVVNGTRR441 pKa = 11.84VWKK444 pKa = 10.43KK445 pKa = 10.14KK446 pKa = 10.65SSDD449 pKa = 3.18GLMTGVPGTTLFDD462 pKa = 3.52TVKK465 pKa = 10.95AIVSWNFCLEE475 pKa = 3.53ASARR479 pKa = 11.84GEE481 pKa = 4.21VNLLNSQDD489 pKa = 3.31VTQWMRR495 pKa = 11.84DD496 pKa = 3.29QCGLVIKK503 pKa = 10.12PGTYY507 pKa = 10.18NPEE510 pKa = 4.12KK511 pKa = 10.79LPGEE515 pKa = 3.98IPNRR519 pKa = 11.84GEE521 pKa = 3.76LCGKK525 pKa = 10.33GKK527 pKa = 10.07FLGMQIMVEE536 pKa = 4.1EE537 pKa = 4.55FEE539 pKa = 4.63GEE541 pKa = 4.55TVFVPTLPEE550 pKa = 3.65EE551 pKa = 4.37EE552 pKa = 4.42MIEE555 pKa = 3.97MLVVQKK561 pKa = 10.79DD562 pKa = 3.51DD563 pKa = 4.3PYY565 pKa = 11.42QKK567 pKa = 9.3TRR569 pKa = 11.84SLTANQRR576 pKa = 11.84LLFDD580 pKa = 4.01RR581 pKa = 11.84MRR583 pKa = 11.84GLMITFGFTNPLVVSTIHH601 pKa = 6.56NIVNTIPPTAVLMEE615 pKa = 4.1TQIKK619 pKa = 9.84GGSSPEE625 pKa = 4.51HH626 pKa = 6.21ITLQDD631 pKa = 3.28FGYY634 pKa = 9.25PDD636 pKa = 3.68SDD638 pKa = 3.38GFPSVEE644 pKa = 3.95FCKK647 pKa = 10.45AVYY650 pKa = 9.98AGCLEE655 pKa = 4.3RR656 pKa = 11.84DD657 pKa = 4.12RR658 pKa = 11.84YY659 pKa = 6.72WTPLFPGLAEE669 pKa = 4.01VFKK672 pKa = 10.5EE673 pKa = 3.56LRR675 pKa = 11.84EE676 pKa = 4.09NGRR679 pKa = 11.84HH680 pKa = 4.53YY681 pKa = 10.79LRR683 pKa = 11.84SVRR686 pKa = 11.84LVTRR690 pKa = 11.84EE691 pKa = 3.81GVEE694 pKa = 3.77QTVVRR699 pKa = 11.84EE700 pKa = 4.05DD701 pKa = 4.14KK702 pKa = 11.07IMEE705 pKa = 4.41TNVLPVALEE714 pKa = 4.06EE715 pKa = 4.28PRR717 pKa = 11.84VPTNLPDD724 pKa = 3.84FNEE727 pKa = 4.11RR728 pKa = 11.84SHH730 pKa = 8.4IVDD733 pKa = 4.68LRR735 pKa = 11.84GATQKK740 pKa = 10.59FLPDD744 pKa = 3.93LGQSIRR750 pKa = 11.84RR751 pKa = 11.84LLDD754 pKa = 3.41EE755 pKa = 5.1YY756 pKa = 10.73GGSMYY761 pKa = 10.76VGEE764 pKa = 3.85ARR766 pKa = 11.84MRR768 pKa = 11.84FGCSSNALMRR778 pKa = 11.84AASKK782 pKa = 10.71YY783 pKa = 10.16GFWIDD788 pKa = 3.49SLYY791 pKa = 11.13DD792 pKa = 3.56DD793 pKa = 5.92GIISKK798 pKa = 9.9FPIQTPIPTIQDD810 pKa = 3.77HH811 pKa = 6.29IRR813 pKa = 11.84INMEE817 pKa = 3.79EE818 pKa = 4.17NKK820 pKa = 10.54SVVDD824 pKa = 3.57KK825 pKa = 10.51GTEE828 pKa = 3.77QRR830 pKa = 11.84LKK832 pKa = 10.72AIEE835 pKa = 3.92QLDD838 pKa = 3.58VSDD841 pKa = 3.94RR842 pKa = 11.84VVPAVAVLTNPEE854 pKa = 4.15EE855 pKa = 4.32MYY857 pKa = 11.13LDD859 pKa = 3.91AGFMAGLNRR868 pKa = 11.84AAISWRR874 pKa = 11.84GDD876 pKa = 3.26TDD878 pKa = 3.75PLQVLGQFLARR889 pKa = 11.84QPSHH893 pKa = 6.91FQVKK897 pKa = 8.45WSSKK901 pKa = 9.05VVTQTPDD908 pKa = 3.3RR909 pKa = 11.84IQAQLKK915 pKa = 10.09VISQTFDD922 pKa = 2.59GSRR925 pKa = 11.84LVGSVIGPNFRR936 pKa = 11.84MAKK939 pKa = 8.83VYY941 pKa = 10.5LVIHH945 pKa = 6.63IMNVFDD951 pKa = 5.24SMLVQKK957 pKa = 10.9GEE959 pKa = 4.17SEE961 pKa = 3.98IFRR964 pKa = 11.84DD965 pKa = 3.73YY966 pKa = 11.44LDD968 pKa = 3.26QRR970 pKa = 11.84KK971 pKa = 10.14SIFSPRR977 pKa = 11.84KK978 pKa = 8.63PLPPLNPGLEE988 pKa = 3.86PWNWVEE994 pKa = 4.57EE995 pKa = 4.4IEE997 pKa = 4.79NTIPPQWEE1005 pKa = 3.99VLQQTRR1011 pKa = 11.84EE1012 pKa = 4.06DD1013 pKa = 3.61PVLRR1017 pKa = 11.84EE1018 pKa = 3.53IVDD1021 pKa = 3.92FVNQRR1026 pKa = 11.84FAFATEE1032 pKa = 4.01SQKK1035 pKa = 10.45VFLYY1039 pKa = 10.89NRR1041 pKa = 11.84TNEE1044 pKa = 3.89LTLVGFTKK1052 pKa = 10.34KK1053 pKa = 10.35PLMIQMLVKK1062 pKa = 10.31EE1063 pKa = 4.88LYY1065 pKa = 10.74ALLKK1069 pKa = 10.23EE1070 pKa = 4.55FKK1072 pKa = 10.55KK1073 pKa = 10.61RR1074 pKa = 11.84DD1075 pKa = 3.52ATEE1078 pKa = 3.58LRR1080 pKa = 11.84RR1081 pKa = 11.84MYY1083 pKa = 10.08EE1084 pKa = 3.74QTGDD1088 pKa = 3.68SSVSDD1093 pKa = 3.31QTIYY1097 pKa = 10.58PPPAAFADD1105 pKa = 3.91AEE1107 pKa = 4.43EE1108 pKa = 4.51EE1109 pKa = 4.64VPVSQQQVSQVVEE1122 pKa = 4.44VYY1124 pKa = 10.55RR1125 pKa = 11.84DD1126 pKa = 3.55AQSAIGDD1133 pKa = 3.8KK1134 pKa = 10.58LRR1136 pKa = 11.84PEE1138 pKa = 4.14TLVVDD1143 pKa = 4.63RR1144 pKa = 11.84EE1145 pKa = 4.49TSSSAAASRR1154 pKa = 11.84AVGEE1158 pKa = 4.34DD1159 pKa = 3.48VNKK1162 pKa = 10.28KK1163 pKa = 6.75SQRR1166 pKa = 11.84NRR1168 pKa = 11.84RR1169 pKa = 11.84THH1171 pKa = 5.27EE1172 pKa = 3.69RR1173 pKa = 11.84EE1174 pKa = 3.5KK1175 pKa = 10.66RR1176 pKa = 11.84QLRR1179 pKa = 11.84EE1180 pKa = 3.72ARR1182 pKa = 11.84QLRR1185 pKa = 11.84SQRR1188 pKa = 11.84VRR1190 pKa = 11.84DD1191 pKa = 3.59TEE1193 pKa = 3.98QIVRR1197 pKa = 11.84NEE1199 pKa = 3.96PSTGRR1204 pKa = 11.84ANRR1207 pKa = 11.84SRR1209 pKa = 11.84KK1210 pKa = 9.98NMTQEE1215 pKa = 3.47MRR1217 pKa = 11.84TKK1219 pKa = 10.78LNRR1222 pKa = 11.84KK1223 pKa = 7.1VLEE1226 pKa = 3.72RR1227 pKa = 11.84RR1228 pKa = 11.84KK1229 pKa = 9.59RR1230 pKa = 11.84QRR1232 pKa = 11.84KK1233 pKa = 8.26EE1234 pKa = 3.52LTEE1237 pKa = 4.36SKK1239 pKa = 10.84SSS1241 pKa = 3.28
MM1 pKa = 7.59DD2 pKa = 4.07TSNPVVSSDD11 pKa = 2.99RR12 pKa = 11.84RR13 pKa = 11.84SLRR16 pKa = 11.84EE17 pKa = 3.85AKK19 pKa = 10.01EE20 pKa = 3.46LAVVKK25 pKa = 10.31KK26 pKa = 10.43RR27 pKa = 11.84QDD29 pKa = 2.95LEE31 pKa = 4.4YY32 pKa = 10.58IMNVAQTSTRR42 pKa = 11.84VILPVGEE49 pKa = 4.21RR50 pKa = 11.84ALPEE54 pKa = 4.34AEE56 pKa = 4.2VKK58 pKa = 10.74SIILNLKK65 pKa = 10.09DD66 pKa = 3.51KK67 pKa = 10.35EE68 pKa = 4.41PTVRR72 pKa = 11.84TDD74 pKa = 3.33SEE76 pKa = 4.49DD77 pKa = 3.52MLDD80 pKa = 3.33EE81 pKa = 4.44TVFQLVAPSMPVCGLLSQDD100 pKa = 4.22GVPLHH105 pKa = 6.73PPPIHH110 pKa = 5.75NAKK113 pKa = 10.25GRR115 pKa = 11.84VTTNRR120 pKa = 11.84RR121 pKa = 11.84FGGGAYY127 pKa = 9.14KK128 pKa = 10.51PLGIRR133 pKa = 11.84PMVEE137 pKa = 3.1ILGVRR142 pKa = 11.84TQEE145 pKa = 3.42ITKK148 pKa = 9.99VYY150 pKa = 8.91THH152 pKa = 6.38GTWQGFSNRR161 pKa = 11.84LAKK164 pKa = 10.46QMGRR168 pKa = 11.84KK169 pKa = 8.06CVSMGMTIRR178 pKa = 11.84NSMPGVQMDD187 pKa = 3.58KK188 pKa = 9.47NTPRR192 pKa = 11.84RR193 pKa = 11.84IFFDD197 pKa = 3.06ILNKK201 pKa = 10.16KK202 pKa = 8.63MPARR206 pKa = 11.84KK207 pKa = 8.83PSGWPSLSDD216 pKa = 3.79PLDD219 pKa = 4.13TILDD223 pKa = 4.16NIKK226 pKa = 8.76ITLDD230 pKa = 3.26ASAGPPYY237 pKa = 9.5WRR239 pKa = 11.84NKK241 pKa = 8.22VEE243 pKa = 5.39CMDD246 pKa = 4.08QILSEE251 pKa = 4.66GIPLLVKK258 pKa = 10.25ALKK261 pKa = 10.19EE262 pKa = 3.87NRR264 pKa = 11.84LEE266 pKa = 3.87QFMRR270 pKa = 11.84EE271 pKa = 3.8EE272 pKa = 4.59PEE274 pKa = 4.24FFLCEE279 pKa = 4.11VKK281 pKa = 10.95NKK283 pKa = 9.89LDD285 pKa = 3.5RR286 pKa = 11.84YY287 pKa = 10.37GVDD290 pKa = 3.51EE291 pKa = 5.75LDD293 pKa = 3.92VKK295 pKa = 10.58CRR297 pKa = 11.84PYY299 pKa = 9.94ICYY302 pKa = 8.37PAHH305 pKa = 6.61CALLFSVLCQRR316 pKa = 11.84FQEE319 pKa = 4.19KK320 pKa = 11.07LEE322 pKa = 4.1IFTQNFEE329 pKa = 4.01SSNAYY334 pKa = 9.93GFSSANGGLKK344 pKa = 10.37KK345 pKa = 10.35LVEE348 pKa = 4.33WMISTPRR355 pKa = 11.84GTSRR359 pKa = 11.84YY360 pKa = 8.91ICYY363 pKa = 10.56GDD365 pKa = 3.49DD366 pKa = 3.32TRR368 pKa = 11.84IVVADD373 pKa = 4.19RR374 pKa = 11.84NGDD377 pKa = 3.3RR378 pKa = 11.84FMVNPDD384 pKa = 3.72FQQMDD389 pKa = 3.96GSVDD393 pKa = 3.29AQTIAWTIDD402 pKa = 3.5FIIKK406 pKa = 10.27KK407 pKa = 10.32LEE409 pKa = 4.15EE410 pKa = 3.97EE411 pKa = 4.63DD412 pKa = 5.01GEE414 pKa = 4.69EE415 pKa = 3.92NCFWRR420 pKa = 11.84AVAKK424 pKa = 9.94LWKK427 pKa = 10.52LMATNPQFVVNGTRR441 pKa = 11.84VWKK444 pKa = 10.43KK445 pKa = 10.14KK446 pKa = 10.65SSDD449 pKa = 3.18GLMTGVPGTTLFDD462 pKa = 3.52TVKK465 pKa = 10.95AIVSWNFCLEE475 pKa = 3.53ASARR479 pKa = 11.84GEE481 pKa = 4.21VNLLNSQDD489 pKa = 3.31VTQWMRR495 pKa = 11.84DD496 pKa = 3.29QCGLVIKK503 pKa = 10.12PGTYY507 pKa = 10.18NPEE510 pKa = 4.12KK511 pKa = 10.79LPGEE515 pKa = 3.98IPNRR519 pKa = 11.84GEE521 pKa = 3.76LCGKK525 pKa = 10.33GKK527 pKa = 10.07FLGMQIMVEE536 pKa = 4.1EE537 pKa = 4.55FEE539 pKa = 4.63GEE541 pKa = 4.55TVFVPTLPEE550 pKa = 3.65EE551 pKa = 4.37EE552 pKa = 4.42MIEE555 pKa = 3.97MLVVQKK561 pKa = 10.79DD562 pKa = 3.51DD563 pKa = 4.3PYY565 pKa = 11.42QKK567 pKa = 9.3TRR569 pKa = 11.84SLTANQRR576 pKa = 11.84LLFDD580 pKa = 4.01RR581 pKa = 11.84MRR583 pKa = 11.84GLMITFGFTNPLVVSTIHH601 pKa = 6.56NIVNTIPPTAVLMEE615 pKa = 4.1TQIKK619 pKa = 9.84GGSSPEE625 pKa = 4.51HH626 pKa = 6.21ITLQDD631 pKa = 3.28FGYY634 pKa = 9.25PDD636 pKa = 3.68SDD638 pKa = 3.38GFPSVEE644 pKa = 3.95FCKK647 pKa = 10.45AVYY650 pKa = 9.98AGCLEE655 pKa = 4.3RR656 pKa = 11.84DD657 pKa = 4.12RR658 pKa = 11.84YY659 pKa = 6.72WTPLFPGLAEE669 pKa = 4.01VFKK672 pKa = 10.5EE673 pKa = 3.56LRR675 pKa = 11.84EE676 pKa = 4.09NGRR679 pKa = 11.84HH680 pKa = 4.53YY681 pKa = 10.79LRR683 pKa = 11.84SVRR686 pKa = 11.84LVTRR690 pKa = 11.84EE691 pKa = 3.81GVEE694 pKa = 3.77QTVVRR699 pKa = 11.84EE700 pKa = 4.05DD701 pKa = 4.14KK702 pKa = 11.07IMEE705 pKa = 4.41TNVLPVALEE714 pKa = 4.06EE715 pKa = 4.28PRR717 pKa = 11.84VPTNLPDD724 pKa = 3.84FNEE727 pKa = 4.11RR728 pKa = 11.84SHH730 pKa = 8.4IVDD733 pKa = 4.68LRR735 pKa = 11.84GATQKK740 pKa = 10.59FLPDD744 pKa = 3.93LGQSIRR750 pKa = 11.84RR751 pKa = 11.84LLDD754 pKa = 3.41EE755 pKa = 5.1YY756 pKa = 10.73GGSMYY761 pKa = 10.76VGEE764 pKa = 3.85ARR766 pKa = 11.84MRR768 pKa = 11.84FGCSSNALMRR778 pKa = 11.84AASKK782 pKa = 10.71YY783 pKa = 10.16GFWIDD788 pKa = 3.49SLYY791 pKa = 11.13DD792 pKa = 3.56DD793 pKa = 5.92GIISKK798 pKa = 9.9FPIQTPIPTIQDD810 pKa = 3.77HH811 pKa = 6.29IRR813 pKa = 11.84INMEE817 pKa = 3.79EE818 pKa = 4.17NKK820 pKa = 10.54SVVDD824 pKa = 3.57KK825 pKa = 10.51GTEE828 pKa = 3.77QRR830 pKa = 11.84LKK832 pKa = 10.72AIEE835 pKa = 3.92QLDD838 pKa = 3.58VSDD841 pKa = 3.94RR842 pKa = 11.84VVPAVAVLTNPEE854 pKa = 4.15EE855 pKa = 4.32MYY857 pKa = 11.13LDD859 pKa = 3.91AGFMAGLNRR868 pKa = 11.84AAISWRR874 pKa = 11.84GDD876 pKa = 3.26TDD878 pKa = 3.75PLQVLGQFLARR889 pKa = 11.84QPSHH893 pKa = 6.91FQVKK897 pKa = 8.45WSSKK901 pKa = 9.05VVTQTPDD908 pKa = 3.3RR909 pKa = 11.84IQAQLKK915 pKa = 10.09VISQTFDD922 pKa = 2.59GSRR925 pKa = 11.84LVGSVIGPNFRR936 pKa = 11.84MAKK939 pKa = 8.83VYY941 pKa = 10.5LVIHH945 pKa = 6.63IMNVFDD951 pKa = 5.24SMLVQKK957 pKa = 10.9GEE959 pKa = 4.17SEE961 pKa = 3.98IFRR964 pKa = 11.84DD965 pKa = 3.73YY966 pKa = 11.44LDD968 pKa = 3.26QRR970 pKa = 11.84KK971 pKa = 10.14SIFSPRR977 pKa = 11.84KK978 pKa = 8.63PLPPLNPGLEE988 pKa = 3.86PWNWVEE994 pKa = 4.57EE995 pKa = 4.4IEE997 pKa = 4.79NTIPPQWEE1005 pKa = 3.99VLQQTRR1011 pKa = 11.84EE1012 pKa = 4.06DD1013 pKa = 3.61PVLRR1017 pKa = 11.84EE1018 pKa = 3.53IVDD1021 pKa = 3.92FVNQRR1026 pKa = 11.84FAFATEE1032 pKa = 4.01SQKK1035 pKa = 10.45VFLYY1039 pKa = 10.89NRR1041 pKa = 11.84TNEE1044 pKa = 3.89LTLVGFTKK1052 pKa = 10.34KK1053 pKa = 10.35PLMIQMLVKK1062 pKa = 10.31EE1063 pKa = 4.88LYY1065 pKa = 10.74ALLKK1069 pKa = 10.23EE1070 pKa = 4.55FKK1072 pKa = 10.55KK1073 pKa = 10.61RR1074 pKa = 11.84DD1075 pKa = 3.52ATEE1078 pKa = 3.58LRR1080 pKa = 11.84RR1081 pKa = 11.84MYY1083 pKa = 10.08EE1084 pKa = 3.74QTGDD1088 pKa = 3.68SSVSDD1093 pKa = 3.31QTIYY1097 pKa = 10.58PPPAAFADD1105 pKa = 3.91AEE1107 pKa = 4.43EE1108 pKa = 4.51EE1109 pKa = 4.64VPVSQQQVSQVVEE1122 pKa = 4.44VYY1124 pKa = 10.55RR1125 pKa = 11.84DD1126 pKa = 3.55AQSAIGDD1133 pKa = 3.8KK1134 pKa = 10.58LRR1136 pKa = 11.84PEE1138 pKa = 4.14TLVVDD1143 pKa = 4.63RR1144 pKa = 11.84EE1145 pKa = 4.49TSSSAAASRR1154 pKa = 11.84AVGEE1158 pKa = 4.34DD1159 pKa = 3.48VNKK1162 pKa = 10.28KK1163 pKa = 6.75SQRR1166 pKa = 11.84NRR1168 pKa = 11.84RR1169 pKa = 11.84THH1171 pKa = 5.27EE1172 pKa = 3.69RR1173 pKa = 11.84EE1174 pKa = 3.5KK1175 pKa = 10.66RR1176 pKa = 11.84QLRR1179 pKa = 11.84EE1180 pKa = 3.72ARR1182 pKa = 11.84QLRR1185 pKa = 11.84SQRR1188 pKa = 11.84VRR1190 pKa = 11.84DD1191 pKa = 3.59TEE1193 pKa = 3.98QIVRR1197 pKa = 11.84NEE1199 pKa = 3.96PSTGRR1204 pKa = 11.84ANRR1207 pKa = 11.84SRR1209 pKa = 11.84KK1210 pKa = 9.98NMTQEE1215 pKa = 3.47MRR1217 pKa = 11.84TKK1219 pKa = 10.78LNRR1222 pKa = 11.84KK1223 pKa = 7.1VLEE1226 pKa = 3.72RR1227 pKa = 11.84RR1228 pKa = 11.84KK1229 pKa = 9.59RR1230 pKa = 11.84QRR1232 pKa = 11.84KK1233 pKa = 8.26EE1234 pKa = 3.52LTEE1237 pKa = 4.36SKK1239 pKa = 10.84SSS1241 pKa = 3.28
Molecular weight: 141.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KHW2|A0A1L3KHW2_9VIRU Putative capsid protein OS=Hubei permutotetra-like virus 6 OX=1923080 PE=4 SV=1
MM1 pKa = 7.51NEE3 pKa = 3.18TMAGVNRR10 pKa = 11.84DD11 pKa = 3.46KK12 pKa = 11.41NSNSSSKK19 pKa = 11.19GNGVKK24 pKa = 10.41KK25 pKa = 10.11PLTPQQVANRR35 pKa = 11.84KK36 pKa = 8.86AKK38 pKa = 8.93NARR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84EE44 pKa = 3.78RR45 pKa = 11.84RR46 pKa = 11.84KK47 pKa = 10.51NKK49 pKa = 9.71LAEE52 pKa = 4.14SQVMGLTPMMFRR64 pKa = 11.84GGEE67 pKa = 3.96AGLNPADD74 pKa = 4.11PRR76 pKa = 11.84SEE78 pKa = 4.02TSVLQMGSTPSGRR91 pKa = 11.84TAALKK96 pKa = 10.11VLHH99 pKa = 7.01PNMEE103 pKa = 4.28ACQMVVKK110 pKa = 10.36FPDD113 pKa = 4.01GAVSTSVSMEE123 pKa = 3.57RR124 pKa = 11.84RR125 pKa = 11.84DD126 pKa = 3.7EE127 pKa = 4.32FEE129 pKa = 4.66IPPFPPTVSTDD140 pKa = 2.64KK141 pKa = 9.98WDD143 pKa = 4.74CIVMHH148 pKa = 6.79LPFLVGRR155 pKa = 11.84QGVIKK160 pKa = 10.06WNSTLTTPSEE170 pKa = 3.97ATLQRR175 pKa = 11.84VVQVLLEE182 pKa = 4.59NIGSPDD188 pKa = 3.02RR189 pKa = 11.84QYY191 pKa = 11.32PLWYY195 pKa = 9.8AYY197 pKa = 8.78TADD200 pKa = 5.47GIPFLISILSSSVLTSGVAYY220 pKa = 10.47SGVSGLSLYY229 pKa = 10.68AKK231 pKa = 9.99SIRR234 pKa = 11.84RR235 pKa = 11.84TCLGVTTDD243 pKa = 4.99LDD245 pKa = 4.8ASDD248 pKa = 5.07LYY250 pKa = 11.67NNGRR254 pKa = 11.84VVSGQWTPDD263 pKa = 2.99VTTARR268 pKa = 11.84HH269 pKa = 5.71DD270 pKa = 3.51TTDD273 pKa = 2.87IVTGFVMHH281 pKa = 6.93TFDD284 pKa = 4.14IYY286 pKa = 11.26QMQVPAVTTSSIVSSDD302 pKa = 3.11EE303 pKa = 4.0FSRR306 pKa = 11.84QAEE309 pKa = 4.39AKK311 pKa = 8.28TGSYY315 pKa = 9.31MPLRR319 pKa = 11.84TCSSTINMTPVQQWRR334 pKa = 11.84NLAIEE339 pKa = 4.43LPGVDD344 pKa = 4.39ADD346 pKa = 4.79TITSSNCNDD355 pKa = 4.07LFLQGWSIGVEE366 pKa = 4.18YY367 pKa = 10.08WSGLSNQANLRR378 pKa = 11.84MKK380 pKa = 10.32VRR382 pKa = 11.84EE383 pKa = 4.13DD384 pKa = 3.63LEE386 pKa = 4.54IVPSPDD392 pKa = 2.97SPYY395 pKa = 11.31SPFATPAYY403 pKa = 8.76PDD405 pKa = 3.68DD406 pKa = 3.53VRR408 pKa = 11.84ARR410 pKa = 11.84AVIQEE415 pKa = 4.22FSRR418 pKa = 11.84KK419 pKa = 8.52QPHH422 pKa = 7.34AYY424 pKa = 7.56PASYY428 pKa = 11.0NEE430 pKa = 3.91NNILLKK436 pKa = 10.81KK437 pKa = 10.14LISAVGEE444 pKa = 4.22VVEE447 pKa = 5.63DD448 pKa = 3.65LGLPILSPLVKK459 pKa = 10.18GVRR462 pKa = 11.84NFLDD466 pKa = 3.5SKK468 pKa = 10.43LWSHH472 pKa = 7.32ISNLFF477 pKa = 3.33
MM1 pKa = 7.51NEE3 pKa = 3.18TMAGVNRR10 pKa = 11.84DD11 pKa = 3.46KK12 pKa = 11.41NSNSSSKK19 pKa = 11.19GNGVKK24 pKa = 10.41KK25 pKa = 10.11PLTPQQVANRR35 pKa = 11.84KK36 pKa = 8.86AKK38 pKa = 8.93NARR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84EE44 pKa = 3.78RR45 pKa = 11.84RR46 pKa = 11.84KK47 pKa = 10.51NKK49 pKa = 9.71LAEE52 pKa = 4.14SQVMGLTPMMFRR64 pKa = 11.84GGEE67 pKa = 3.96AGLNPADD74 pKa = 4.11PRR76 pKa = 11.84SEE78 pKa = 4.02TSVLQMGSTPSGRR91 pKa = 11.84TAALKK96 pKa = 10.11VLHH99 pKa = 7.01PNMEE103 pKa = 4.28ACQMVVKK110 pKa = 10.36FPDD113 pKa = 4.01GAVSTSVSMEE123 pKa = 3.57RR124 pKa = 11.84RR125 pKa = 11.84DD126 pKa = 3.7EE127 pKa = 4.32FEE129 pKa = 4.66IPPFPPTVSTDD140 pKa = 2.64KK141 pKa = 9.98WDD143 pKa = 4.74CIVMHH148 pKa = 6.79LPFLVGRR155 pKa = 11.84QGVIKK160 pKa = 10.06WNSTLTTPSEE170 pKa = 3.97ATLQRR175 pKa = 11.84VVQVLLEE182 pKa = 4.59NIGSPDD188 pKa = 3.02RR189 pKa = 11.84QYY191 pKa = 11.32PLWYY195 pKa = 9.8AYY197 pKa = 8.78TADD200 pKa = 5.47GIPFLISILSSSVLTSGVAYY220 pKa = 10.47SGVSGLSLYY229 pKa = 10.68AKK231 pKa = 9.99SIRR234 pKa = 11.84RR235 pKa = 11.84TCLGVTTDD243 pKa = 4.99LDD245 pKa = 4.8ASDD248 pKa = 5.07LYY250 pKa = 11.67NNGRR254 pKa = 11.84VVSGQWTPDD263 pKa = 2.99VTTARR268 pKa = 11.84HH269 pKa = 5.71DD270 pKa = 3.51TTDD273 pKa = 2.87IVTGFVMHH281 pKa = 6.93TFDD284 pKa = 4.14IYY286 pKa = 11.26QMQVPAVTTSSIVSSDD302 pKa = 3.11EE303 pKa = 4.0FSRR306 pKa = 11.84QAEE309 pKa = 4.39AKK311 pKa = 8.28TGSYY315 pKa = 9.31MPLRR319 pKa = 11.84TCSSTINMTPVQQWRR334 pKa = 11.84NLAIEE339 pKa = 4.43LPGVDD344 pKa = 4.39ADD346 pKa = 4.79TITSSNCNDD355 pKa = 4.07LFLQGWSIGVEE366 pKa = 4.18YY367 pKa = 10.08WSGLSNQANLRR378 pKa = 11.84MKK380 pKa = 10.32VRR382 pKa = 11.84EE383 pKa = 4.13DD384 pKa = 3.63LEE386 pKa = 4.54IVPSPDD392 pKa = 2.97SPYY395 pKa = 11.31SPFATPAYY403 pKa = 8.76PDD405 pKa = 3.68DD406 pKa = 3.53VRR408 pKa = 11.84ARR410 pKa = 11.84AVIQEE415 pKa = 4.22FSRR418 pKa = 11.84KK419 pKa = 8.52QPHH422 pKa = 7.34AYY424 pKa = 7.56PASYY428 pKa = 11.0NEE430 pKa = 3.91NNILLKK436 pKa = 10.81KK437 pKa = 10.14LISAVGEE444 pKa = 4.22VVEE447 pKa = 5.63DD448 pKa = 3.65LGLPILSPLVKK459 pKa = 10.18GVRR462 pKa = 11.84NFLDD466 pKa = 3.5SKK468 pKa = 10.43LWSHH472 pKa = 7.32ISNLFF477 pKa = 3.33
Molecular weight: 52.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1718 |
477 |
1241 |
859.0 |
97.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.471 ± 0.51 | 1.222 ± 0.086 |
5.471 ± 0.114 | 6.752 ± 1.166 |
3.783 ± 0.421 | 5.879 ± 0.1 |
1.048 ± 0.104 | 4.889 ± 0.242 |
5.413 ± 0.606 | 8.44 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.26 ± 0.057 | 4.54 ± 0.348 |
6.112 ± 0.296 | 4.773 ± 0.392 |
7.276 ± 0.698 | 7.334 ± 1.563 |
6.17 ± 0.475 | 8.324 ± 0.135 |
1.455 ± 0.11 | 2.386 ± 0.168 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
