
Capybara microvirus Cap1_SP_145
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
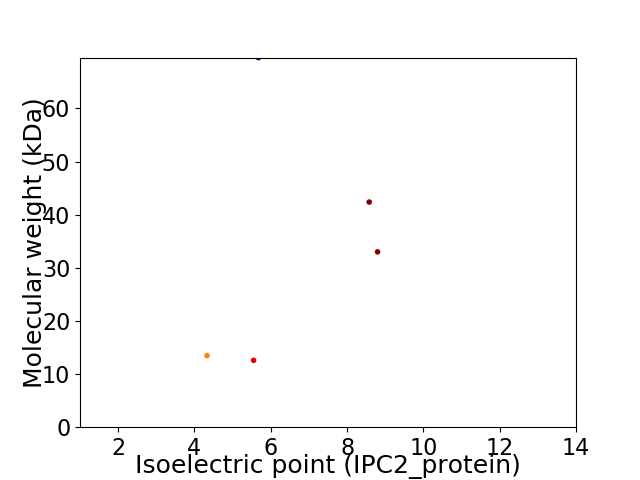
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W7X8|A0A4P8W7X8_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_145 OX=2585390 PE=4 SV=1
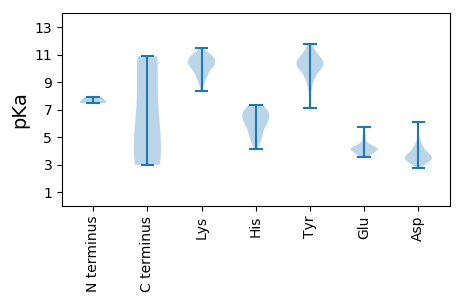
MM1 pKa = 7.83KK2 pKa = 10.49YY3 pKa = 10.56LEE5 pKa = 5.05RR6 pKa = 11.84DD7 pKa = 3.17NKK9 pKa = 10.97SIVTVTEE16 pKa = 3.85YY17 pKa = 10.48MSLAEE22 pKa = 4.46CIAYY26 pKa = 9.02YY27 pKa = 10.63EE28 pKa = 4.47RR29 pKa = 11.84TNEE32 pKa = 4.08LPPIGEE38 pKa = 4.47TKK40 pKa = 10.81GEE42 pKa = 4.08YY43 pKa = 9.98DD44 pKa = 3.68DD45 pKa = 6.11SITISEE51 pKa = 4.31AEE53 pKa = 3.74ARR55 pKa = 11.84TINVAKK61 pKa = 10.38LYY63 pKa = 10.81KK64 pKa = 10.21FDD66 pKa = 4.95DD67 pKa = 4.24DD68 pKa = 4.54RR69 pKa = 11.84VSDD72 pKa = 3.82MDD74 pKa = 3.48ISDD77 pKa = 3.81MRR79 pKa = 11.84NSSSEE84 pKa = 4.15SYY86 pKa = 10.45ILEE89 pKa = 4.09ASAEE93 pKa = 4.03KK94 pKa = 10.5SAEE97 pKa = 3.8KK98 pKa = 10.31SAEE101 pKa = 3.63ISVEE105 pKa = 4.11NISVEE110 pKa = 4.2QVSPQATAEE119 pKa = 4.24KK120 pKa = 10.68
MM1 pKa = 7.83KK2 pKa = 10.49YY3 pKa = 10.56LEE5 pKa = 5.05RR6 pKa = 11.84DD7 pKa = 3.17NKK9 pKa = 10.97SIVTVTEE16 pKa = 3.85YY17 pKa = 10.48MSLAEE22 pKa = 4.46CIAYY26 pKa = 9.02YY27 pKa = 10.63EE28 pKa = 4.47RR29 pKa = 11.84TNEE32 pKa = 4.08LPPIGEE38 pKa = 4.47TKK40 pKa = 10.81GEE42 pKa = 4.08YY43 pKa = 9.98DD44 pKa = 3.68DD45 pKa = 6.11SITISEE51 pKa = 4.31AEE53 pKa = 3.74ARR55 pKa = 11.84TINVAKK61 pKa = 10.38LYY63 pKa = 10.81KK64 pKa = 10.21FDD66 pKa = 4.95DD67 pKa = 4.24DD68 pKa = 4.54RR69 pKa = 11.84VSDD72 pKa = 3.82MDD74 pKa = 3.48ISDD77 pKa = 3.81MRR79 pKa = 11.84NSSSEE84 pKa = 4.15SYY86 pKa = 10.45ILEE89 pKa = 4.09ASAEE93 pKa = 4.03KK94 pKa = 10.5SAEE97 pKa = 3.8KK98 pKa = 10.31SAEE101 pKa = 3.63ISVEE105 pKa = 4.11NISVEE110 pKa = 4.2QVSPQATAEE119 pKa = 4.24KK120 pKa = 10.68
Molecular weight: 13.48 kDa
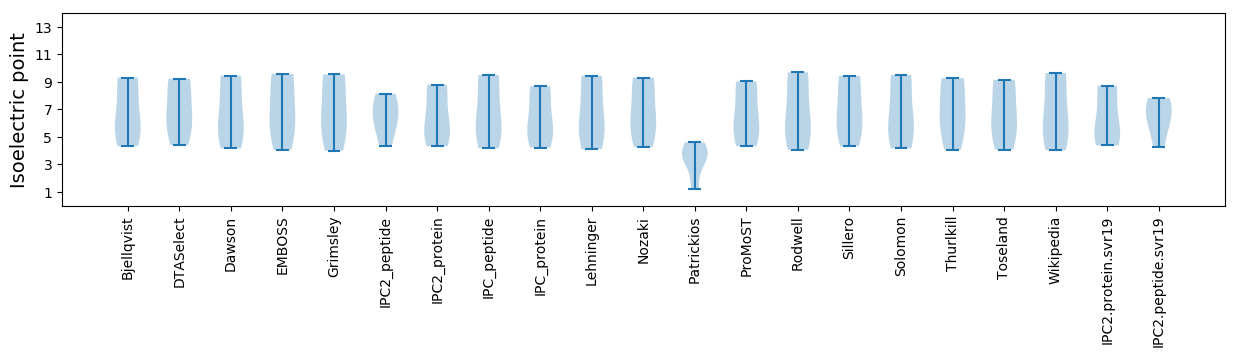
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W5G0|A0A4P8W5G0_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_145 OX=2585390 PE=4 SV=1
MM1 pKa = 7.53GSGLHH6 pKa = 6.05RR7 pKa = 11.84VNTVPCGKK15 pKa = 10.05CVACRR20 pKa = 11.84SNKK23 pKa = 9.11RR24 pKa = 11.84EE25 pKa = 3.69EE26 pKa = 4.09WASRR30 pKa = 11.84LAMEE34 pKa = 5.34YY35 pKa = 9.22YY36 pKa = 7.87TTFKK40 pKa = 10.78KK41 pKa = 10.77GFEE44 pKa = 4.32TFFITLTFNDD54 pKa = 4.37EE55 pKa = 4.08NLQNPSLDD63 pKa = 3.44KK64 pKa = 10.98RR65 pKa = 11.84DD66 pKa = 3.23IQLFNKK72 pKa = 9.7KK73 pKa = 8.87FRR75 pKa = 11.84HH76 pKa = 5.05YY77 pKa = 10.94LSYY80 pKa = 10.95KK81 pKa = 9.75YY82 pKa = 8.95FFCGEE87 pKa = 4.11YY88 pKa = 10.56GDD90 pKa = 4.28KK91 pKa = 10.39FDD93 pKa = 4.43RR94 pKa = 11.84KK95 pKa = 9.63HH96 pKa = 4.14YY97 pKa = 10.02HH98 pKa = 6.86ALIFCNTVVTPEE110 pKa = 3.92YY111 pKa = 9.27FTRR114 pKa = 11.84SLEE117 pKa = 4.14KK118 pKa = 9.13CWPYY122 pKa = 11.54GFVKK126 pKa = 10.64ADD128 pKa = 2.94NFTYY132 pKa = 10.74ARR134 pKa = 11.84ARR136 pKa = 11.84YY137 pKa = 7.08VAKK140 pKa = 10.73YY141 pKa = 10.24ALKK144 pKa = 10.43DD145 pKa = 4.13DD146 pKa = 3.92NWSDD150 pKa = 3.95TNLSPPFALMSTKK163 pKa = 10.14PAICYY168 pKa = 10.14DD169 pKa = 3.87YY170 pKa = 11.74VKK172 pKa = 10.57LFHH175 pKa = 6.12NQIYY179 pKa = 10.37DD180 pKa = 3.78GQSLFYY186 pKa = 9.53CDD188 pKa = 2.75SVGYY192 pKa = 9.99HH193 pKa = 6.36RR194 pKa = 11.84LPRR197 pKa = 11.84YY198 pKa = 9.07VKK200 pKa = 10.07EE201 pKa = 4.48KK202 pKa = 10.11IFSKK206 pKa = 10.01IWLRR210 pKa = 11.84EE211 pKa = 3.67QNDD214 pKa = 3.75AYY216 pKa = 11.41SRR218 pKa = 11.84LEE220 pKa = 3.96FNKK223 pKa = 10.08EE224 pKa = 3.78LYY226 pKa = 10.15VLRR229 pKa = 11.84NLQLLHH235 pKa = 6.7GGNHH239 pKa = 5.91NAVNSAYY246 pKa = 10.01RR247 pKa = 11.84AYY249 pKa = 9.83MYY251 pKa = 10.37QKK253 pKa = 10.41YY254 pKa = 9.9KK255 pKa = 10.78SPEE258 pKa = 3.55VNYY261 pKa = 10.02FSNKK265 pKa = 9.25YY266 pKa = 9.4IYY268 pKa = 10.75SRR270 pKa = 11.84FKK272 pKa = 11.5NNFF275 pKa = 2.99
MM1 pKa = 7.53GSGLHH6 pKa = 6.05RR7 pKa = 11.84VNTVPCGKK15 pKa = 10.05CVACRR20 pKa = 11.84SNKK23 pKa = 9.11RR24 pKa = 11.84EE25 pKa = 3.69EE26 pKa = 4.09WASRR30 pKa = 11.84LAMEE34 pKa = 5.34YY35 pKa = 9.22YY36 pKa = 7.87TTFKK40 pKa = 10.78KK41 pKa = 10.77GFEE44 pKa = 4.32TFFITLTFNDD54 pKa = 4.37EE55 pKa = 4.08NLQNPSLDD63 pKa = 3.44KK64 pKa = 10.98RR65 pKa = 11.84DD66 pKa = 3.23IQLFNKK72 pKa = 9.7KK73 pKa = 8.87FRR75 pKa = 11.84HH76 pKa = 5.05YY77 pKa = 10.94LSYY80 pKa = 10.95KK81 pKa = 9.75YY82 pKa = 8.95FFCGEE87 pKa = 4.11YY88 pKa = 10.56GDD90 pKa = 4.28KK91 pKa = 10.39FDD93 pKa = 4.43RR94 pKa = 11.84KK95 pKa = 9.63HH96 pKa = 4.14YY97 pKa = 10.02HH98 pKa = 6.86ALIFCNTVVTPEE110 pKa = 3.92YY111 pKa = 9.27FTRR114 pKa = 11.84SLEE117 pKa = 4.14KK118 pKa = 9.13CWPYY122 pKa = 11.54GFVKK126 pKa = 10.64ADD128 pKa = 2.94NFTYY132 pKa = 10.74ARR134 pKa = 11.84ARR136 pKa = 11.84YY137 pKa = 7.08VAKK140 pKa = 10.73YY141 pKa = 10.24ALKK144 pKa = 10.43DD145 pKa = 4.13DD146 pKa = 3.92NWSDD150 pKa = 3.95TNLSPPFALMSTKK163 pKa = 10.14PAICYY168 pKa = 10.14DD169 pKa = 3.87YY170 pKa = 11.74VKK172 pKa = 10.57LFHH175 pKa = 6.12NQIYY179 pKa = 10.37DD180 pKa = 3.78GQSLFYY186 pKa = 9.53CDD188 pKa = 2.75SVGYY192 pKa = 9.99HH193 pKa = 6.36RR194 pKa = 11.84LPRR197 pKa = 11.84YY198 pKa = 9.07VKK200 pKa = 10.07EE201 pKa = 4.48KK202 pKa = 10.11IFSKK206 pKa = 10.01IWLRR210 pKa = 11.84EE211 pKa = 3.67QNDD214 pKa = 3.75AYY216 pKa = 11.41SRR218 pKa = 11.84LEE220 pKa = 3.96FNKK223 pKa = 10.08EE224 pKa = 3.78LYY226 pKa = 10.15VLRR229 pKa = 11.84NLQLLHH235 pKa = 6.7GGNHH239 pKa = 5.91NAVNSAYY246 pKa = 10.01RR247 pKa = 11.84AYY249 pKa = 9.83MYY251 pKa = 10.37QKK253 pKa = 10.41YY254 pKa = 9.9KK255 pKa = 10.78SPEE258 pKa = 3.55VNYY261 pKa = 10.02FSNKK265 pKa = 9.25YY266 pKa = 9.4IYY268 pKa = 10.75SRR270 pKa = 11.84FKK272 pKa = 11.5NNFF275 pKa = 2.99
Molecular weight: 33.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1482 |
106 |
604 |
296.4 |
34.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.478 ± 0.56 | 1.282 ± 0.429 |
4.858 ± 0.775 | 6.005 ± 1.09 |
5.398 ± 0.916 | 4.858 ± 0.625 |
1.619 ± 0.554 | 5.87 ± 0.689 |
6.883 ± 0.554 | 9.109 ± 1.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.092 ± 0.173 | 7.152 ± 0.949 |
3.509 ± 0.411 | 4.926 ± 1.167 |
3.914 ± 0.529 | 8.367 ± 0.683 |
5.466 ± 0.365 | 3.914 ± 0.562 |
1.552 ± 0.224 | 6.748 ± 0.861 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
