
Cervid alphaherpesvirus 2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Varicellovirus
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
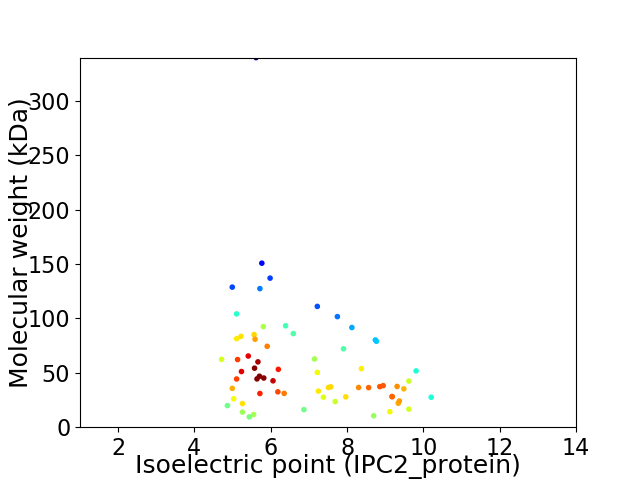
Virtual 2D-PAGE plot for 68 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

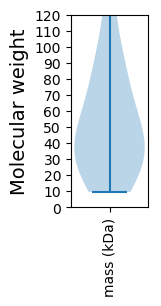
Protein with the lowest isoelectric point:
>tr|A0A455JQ30|A0A455JQ30_9ALPH DNA primase OS=Cervid alphaherpesvirus 2 OX=365327 GN=UL52 PE=3 SV=1
MM1 pKa = 7.77CAPPPPPPPPPSRR14 pKa = 11.84WLPLLLLLWPLLWPPLGAGAEE35 pKa = 4.16PGPGALARR43 pKa = 11.84PVGVIVARR51 pKa = 11.84AGAPVFLPGDD61 pKa = 3.58AAVPRR66 pKa = 11.84PGPAPGPDD74 pKa = 2.86ARR76 pKa = 11.84AVRR79 pKa = 11.84GWSRR83 pKa = 11.84LASACPPPGPATEE96 pKa = 4.1VCIDD100 pKa = 3.44GHH102 pKa = 5.29QCFADD107 pKa = 3.76VVVDD111 pKa = 4.31AACLPTVSLEE121 pKa = 4.01PLAVAEE127 pKa = 4.16LSGGGGAGRR136 pKa = 11.84LADD139 pKa = 3.73LAEE142 pKa = 4.02RR143 pKa = 11.84RR144 pKa = 11.84VFAQLDD150 pKa = 3.85YY151 pKa = 11.59NEE153 pKa = 4.26TGVWIPNATAADD165 pKa = 3.46AGVYY169 pKa = 9.57FLYY172 pKa = 10.54DD173 pKa = 3.22RR174 pKa = 11.84RR175 pKa = 11.84LGEE178 pKa = 4.19AGGEE182 pKa = 4.17EE183 pKa = 4.33THH185 pKa = 6.58SALALRR191 pKa = 11.84VEE193 pKa = 4.29ADD195 pKa = 3.92EE196 pKa = 5.03IEE198 pKa = 4.32GDD200 pKa = 3.76EE201 pKa = 4.6DD202 pKa = 3.58EE203 pKa = 4.38EE204 pKa = 4.25AARR207 pKa = 11.84GRR209 pKa = 11.84STAAPRR215 pKa = 11.84TPAPRR220 pKa = 11.84TPAAPSPAPARR231 pKa = 11.84HH232 pKa = 5.59GARR235 pKa = 11.84FRR237 pKa = 11.84VLPYY241 pKa = 9.52HH242 pKa = 5.24SHH244 pKa = 6.29VYY246 pKa = 9.57TPGDD250 pKa = 3.7SFQLAVRR257 pKa = 11.84LQSEE261 pKa = 4.88FFDD264 pKa = 3.73EE265 pKa = 4.8TPFAAAIDD273 pKa = 4.11WYY275 pKa = 9.92FLRR278 pKa = 11.84PAADD282 pKa = 3.52CALVRR287 pKa = 11.84LYY289 pKa = 7.34EE290 pKa = 4.0TCIFHH295 pKa = 7.43PEE297 pKa = 3.75APACLHH303 pKa = 6.34PVDD306 pKa = 4.85ARR308 pKa = 11.84CSFASPYY315 pKa = 10.14RR316 pKa = 11.84SEE318 pKa = 4.15TVYY321 pKa = 10.85GRR323 pKa = 11.84LYY325 pKa = 9.87PRR327 pKa = 11.84CDD329 pKa = 3.19PAAGARR335 pKa = 11.84WPRR338 pKa = 11.84EE339 pKa = 4.11CEE341 pKa = 3.97GASYY345 pKa = 7.92TAPGAHH351 pKa = 6.72ARR353 pKa = 11.84PAEE356 pKa = 4.24NGADD360 pKa = 3.84LVFADD365 pKa = 5.18APAAAAGLYY374 pKa = 10.31VFVLQYY380 pKa = 10.55NGHH383 pKa = 5.37VEE385 pKa = 3.7AWDD388 pKa = 3.77YY389 pKa = 11.88SLVATSDD396 pKa = 3.38RR397 pKa = 11.84LVRR400 pKa = 11.84AVVDD404 pKa = 3.49HH405 pKa = 6.23TRR407 pKa = 11.84PEE409 pKa = 4.04AAEE412 pKa = 4.55GPGPSAAPGGGEE424 pKa = 4.17PASAPAPARR433 pKa = 11.84RR434 pKa = 11.84FAPWLVALGGVLALVALGGLAALGVWGCAQRR465 pKa = 11.84AARR468 pKa = 11.84RR469 pKa = 11.84RR470 pKa = 11.84TYY472 pKa = 10.78DD473 pKa = 2.73ILNPFGSAYY482 pKa = 9.76TSLPTNDD489 pKa = 3.89PFEE492 pKa = 5.04SSGEE496 pKa = 4.11EE497 pKa = 3.74EE498 pKa = 4.51GGEE501 pKa = 4.28DD502 pKa = 4.65FPPPPGDD509 pKa = 3.43EE510 pKa = 3.88TDD512 pKa = 4.65SFDD515 pKa = 3.89EE516 pKa = 5.38DD517 pKa = 3.92SDD519 pKa = 4.2EE520 pKa = 4.58EE521 pKa = 4.11PSAPYY526 pKa = 10.18EE527 pKa = 4.12LADD530 pKa = 4.63APYY533 pKa = 9.81EE534 pKa = 4.05LADD537 pKa = 4.23AEE539 pKa = 4.43PAGLARR545 pKa = 11.84TPAGGPRR552 pKa = 11.84ASRR555 pKa = 11.84SGFKK559 pKa = 9.97VWFRR563 pKa = 11.84DD564 pKa = 3.65PVEE567 pKa = 4.55EE568 pKa = 4.29EE569 pKa = 4.07PGSPRR574 pKa = 11.84LPAAPDD580 pKa = 3.45YY581 pKa = 9.92TAVAARR587 pKa = 11.84LKK589 pKa = 10.85SILRR593 pKa = 3.86
MM1 pKa = 7.77CAPPPPPPPPPSRR14 pKa = 11.84WLPLLLLLWPLLWPPLGAGAEE35 pKa = 4.16PGPGALARR43 pKa = 11.84PVGVIVARR51 pKa = 11.84AGAPVFLPGDD61 pKa = 3.58AAVPRR66 pKa = 11.84PGPAPGPDD74 pKa = 2.86ARR76 pKa = 11.84AVRR79 pKa = 11.84GWSRR83 pKa = 11.84LASACPPPGPATEE96 pKa = 4.1VCIDD100 pKa = 3.44GHH102 pKa = 5.29QCFADD107 pKa = 3.76VVVDD111 pKa = 4.31AACLPTVSLEE121 pKa = 4.01PLAVAEE127 pKa = 4.16LSGGGGAGRR136 pKa = 11.84LADD139 pKa = 3.73LAEE142 pKa = 4.02RR143 pKa = 11.84RR144 pKa = 11.84VFAQLDD150 pKa = 3.85YY151 pKa = 11.59NEE153 pKa = 4.26TGVWIPNATAADD165 pKa = 3.46AGVYY169 pKa = 9.57FLYY172 pKa = 10.54DD173 pKa = 3.22RR174 pKa = 11.84RR175 pKa = 11.84LGEE178 pKa = 4.19AGGEE182 pKa = 4.17EE183 pKa = 4.33THH185 pKa = 6.58SALALRR191 pKa = 11.84VEE193 pKa = 4.29ADD195 pKa = 3.92EE196 pKa = 5.03IEE198 pKa = 4.32GDD200 pKa = 3.76EE201 pKa = 4.6DD202 pKa = 3.58EE203 pKa = 4.38EE204 pKa = 4.25AARR207 pKa = 11.84GRR209 pKa = 11.84STAAPRR215 pKa = 11.84TPAPRR220 pKa = 11.84TPAAPSPAPARR231 pKa = 11.84HH232 pKa = 5.59GARR235 pKa = 11.84FRR237 pKa = 11.84VLPYY241 pKa = 9.52HH242 pKa = 5.24SHH244 pKa = 6.29VYY246 pKa = 9.57TPGDD250 pKa = 3.7SFQLAVRR257 pKa = 11.84LQSEE261 pKa = 4.88FFDD264 pKa = 3.73EE265 pKa = 4.8TPFAAAIDD273 pKa = 4.11WYY275 pKa = 9.92FLRR278 pKa = 11.84PAADD282 pKa = 3.52CALVRR287 pKa = 11.84LYY289 pKa = 7.34EE290 pKa = 4.0TCIFHH295 pKa = 7.43PEE297 pKa = 3.75APACLHH303 pKa = 6.34PVDD306 pKa = 4.85ARR308 pKa = 11.84CSFASPYY315 pKa = 10.14RR316 pKa = 11.84SEE318 pKa = 4.15TVYY321 pKa = 10.85GRR323 pKa = 11.84LYY325 pKa = 9.87PRR327 pKa = 11.84CDD329 pKa = 3.19PAAGARR335 pKa = 11.84WPRR338 pKa = 11.84EE339 pKa = 4.11CEE341 pKa = 3.97GASYY345 pKa = 7.92TAPGAHH351 pKa = 6.72ARR353 pKa = 11.84PAEE356 pKa = 4.24NGADD360 pKa = 3.84LVFADD365 pKa = 5.18APAAAAGLYY374 pKa = 10.31VFVLQYY380 pKa = 10.55NGHH383 pKa = 5.37VEE385 pKa = 3.7AWDD388 pKa = 3.77YY389 pKa = 11.88SLVATSDD396 pKa = 3.38RR397 pKa = 11.84LVRR400 pKa = 11.84AVVDD404 pKa = 3.49HH405 pKa = 6.23TRR407 pKa = 11.84PEE409 pKa = 4.04AAEE412 pKa = 4.55GPGPSAAPGGGEE424 pKa = 4.17PASAPAPARR433 pKa = 11.84RR434 pKa = 11.84FAPWLVALGGVLALVALGGLAALGVWGCAQRR465 pKa = 11.84AARR468 pKa = 11.84RR469 pKa = 11.84RR470 pKa = 11.84TYY472 pKa = 10.78DD473 pKa = 2.73ILNPFGSAYY482 pKa = 9.76TSLPTNDD489 pKa = 3.89PFEE492 pKa = 5.04SSGEE496 pKa = 4.11EE497 pKa = 3.74EE498 pKa = 4.51GGEE501 pKa = 4.28DD502 pKa = 4.65FPPPPGDD509 pKa = 3.43EE510 pKa = 3.88TDD512 pKa = 4.65SFDD515 pKa = 3.89EE516 pKa = 5.38DD517 pKa = 3.92SDD519 pKa = 4.2EE520 pKa = 4.58EE521 pKa = 4.11PSAPYY526 pKa = 10.18EE527 pKa = 4.12LADD530 pKa = 4.63APYY533 pKa = 9.81EE534 pKa = 4.05LADD537 pKa = 4.23AEE539 pKa = 4.43PAGLARR545 pKa = 11.84TPAGGPRR552 pKa = 11.84ASRR555 pKa = 11.84SGFKK559 pKa = 9.97VWFRR563 pKa = 11.84DD564 pKa = 3.65PVEE567 pKa = 4.55EE568 pKa = 4.29EE569 pKa = 4.07PGSPRR574 pKa = 11.84LPAAPDD580 pKa = 3.45YY581 pKa = 9.92TAVAARR587 pKa = 11.84LKK589 pKa = 10.85SILRR593 pKa = 3.86
Molecular weight: 62.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A455JQ96|A0A455JQ96_9ALPH Envelope glycoprotein G OS=Cervid alphaherpesvirus 2 OX=365327 GN=US4 PE=4 SV=1
MM1 pKa = 7.6ALARR5 pKa = 11.84TQPPRR10 pKa = 11.84AAGGVFGEE18 pKa = 4.22ARR20 pKa = 11.84VATVADD26 pKa = 4.15YY27 pKa = 11.21KK28 pKa = 11.02RR29 pKa = 11.84LLATNRR35 pKa = 11.84AAAAKK40 pKa = 9.78LRR42 pKa = 11.84AAAGGGEE49 pKa = 4.1AAPDD53 pKa = 3.36EE54 pKa = 4.63RR55 pKa = 11.84GRR57 pKa = 11.84AQTGAGRR64 pKa = 11.84PRR66 pKa = 11.84GGARR70 pKa = 11.84GRR72 pKa = 11.84AAEE75 pKa = 4.27DD76 pKa = 3.41APARR80 pKa = 11.84GARR83 pKa = 11.84RR84 pKa = 11.84PSGRR88 pKa = 11.84AARR91 pKa = 11.84RR92 pKa = 11.84PSGRR96 pKa = 11.84ARR98 pKa = 11.84PAPEE102 pKa = 4.85DD103 pKa = 3.85EE104 pKa = 4.76YY105 pKa = 11.49GDD107 pKa = 3.94EE108 pKa = 4.65YY109 pKa = 11.07EE110 pKa = 5.89DD111 pKa = 3.84EE112 pKa = 4.45YY113 pKa = 11.18EE114 pKa = 4.34YY115 pKa = 11.61GDD117 pKa = 4.11AEE119 pKa = 4.42EE120 pKa = 5.56GPTPAEE126 pKa = 3.69VRR128 pKa = 11.84AARR131 pKa = 11.84LYY133 pKa = 10.75AAAGPARR140 pKa = 11.84ARR142 pKa = 11.84RR143 pKa = 11.84PAPAGEE149 pKa = 4.43FEE151 pKa = 4.15RR152 pKa = 11.84QTRR155 pKa = 11.84AGTRR159 pKa = 3.25
MM1 pKa = 7.6ALARR5 pKa = 11.84TQPPRR10 pKa = 11.84AAGGVFGEE18 pKa = 4.22ARR20 pKa = 11.84VATVADD26 pKa = 4.15YY27 pKa = 11.21KK28 pKa = 11.02RR29 pKa = 11.84LLATNRR35 pKa = 11.84AAAAKK40 pKa = 9.78LRR42 pKa = 11.84AAAGGGEE49 pKa = 4.1AAPDD53 pKa = 3.36EE54 pKa = 4.63RR55 pKa = 11.84GRR57 pKa = 11.84AQTGAGRR64 pKa = 11.84PRR66 pKa = 11.84GGARR70 pKa = 11.84GRR72 pKa = 11.84AAEE75 pKa = 4.27DD76 pKa = 3.41APARR80 pKa = 11.84GARR83 pKa = 11.84RR84 pKa = 11.84PSGRR88 pKa = 11.84AARR91 pKa = 11.84RR92 pKa = 11.84PSGRR96 pKa = 11.84ARR98 pKa = 11.84PAPEE102 pKa = 4.85DD103 pKa = 3.85EE104 pKa = 4.76YY105 pKa = 11.49GDD107 pKa = 3.94EE108 pKa = 4.65YY109 pKa = 11.07EE110 pKa = 5.89DD111 pKa = 3.84EE112 pKa = 4.45YY113 pKa = 11.18EE114 pKa = 4.34YY115 pKa = 11.61GDD117 pKa = 4.11AEE119 pKa = 4.42EE120 pKa = 5.56GPTPAEE126 pKa = 3.69VRR128 pKa = 11.84AARR131 pKa = 11.84LYY133 pKa = 10.75AAAGPARR140 pKa = 11.84ARR142 pKa = 11.84RR143 pKa = 11.84PAPAGEE149 pKa = 4.43FEE151 pKa = 4.15RR152 pKa = 11.84QTRR155 pKa = 11.84AGTRR159 pKa = 3.25
Molecular weight: 16.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
37385 |
88 |
3326 |
549.8 |
57.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
21.391 ± 0.791 | 1.752 ± 0.124 |
4.836 ± 0.129 | 6.144 ± 0.193 |
3.135 ± 0.183 | 8.527 ± 0.206 |
1.765 ± 0.101 | 1.418 ± 0.108 |
1.062 ± 0.124 | 10.304 ± 0.206 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.287 ± 0.114 | 1.412 ± 0.15 |
8.083 ± 0.449 | 2.124 ± 0.148 |
9.739 ± 0.248 | 4.307 ± 0.164 |
3.17 ± 0.162 | 6.275 ± 0.192 |
0.995 ± 0.063 | 2.274 ± 0.19 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
