
Hubei partiti-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
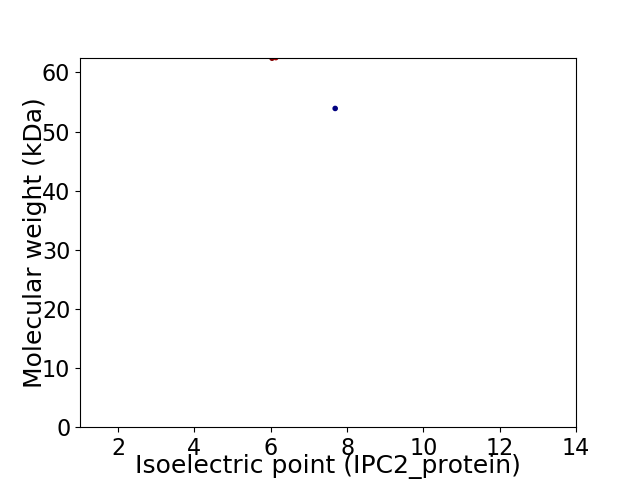
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLP6|A0A1L3KLP6_9VIRU RdRp OS=Hubei partiti-like virus 11 OX=1923017 PE=4 SV=1
MM1 pKa = 7.86EE2 pKa = 4.69YY3 pKa = 10.33LGKK6 pKa = 10.51AEE8 pKa = 4.47GFNMDD13 pKa = 4.36LRR15 pKa = 11.84PPRR18 pKa = 11.84MDD20 pKa = 5.92GITCGYY26 pKa = 8.48MNDD29 pKa = 3.84WFGDD33 pKa = 3.61AYY35 pKa = 11.26VDD37 pKa = 3.94VVMQTYY43 pKa = 9.99HH44 pKa = 7.24RR45 pKa = 11.84SQPSLEE51 pKa = 4.01LLKK54 pKa = 10.65EE55 pKa = 3.72DD56 pKa = 4.28FMRR59 pKa = 11.84YY60 pKa = 8.31DD61 pKa = 3.34RR62 pKa = 11.84KK63 pKa = 10.51YY64 pKa = 8.62VAGTVRR70 pKa = 11.84EE71 pKa = 4.08EE72 pKa = 3.48QSYY75 pKa = 11.03RR76 pKa = 11.84MVLQSVRR83 pKa = 11.84DD84 pKa = 3.67QFVLGEE90 pKa = 4.04KK91 pKa = 10.38LIPLTLGAVFEE102 pKa = 4.92SNSMTTDD109 pKa = 3.46KK110 pKa = 11.42SPGLPWIQRR119 pKa = 11.84GYY121 pKa = 8.11KK122 pKa = 9.04TKK124 pKa = 10.95GDD126 pKa = 3.35VFACHH131 pKa = 6.0EE132 pKa = 4.18ARR134 pKa = 11.84SEE136 pKa = 4.74IFRR139 pKa = 11.84TWDD142 pKa = 3.55RR143 pKa = 11.84IGNGRR148 pKa = 11.84GATLPDD154 pKa = 3.02TCAFMRR160 pKa = 11.84VQLAKK165 pKa = 10.3EE166 pKa = 4.22DD167 pKa = 3.75KK168 pKa = 10.22QKK170 pKa = 10.44IRR172 pKa = 11.84AVWGTPTDD180 pKa = 4.05VIAEE184 pKa = 4.22EE185 pKa = 4.29ARR187 pKa = 11.84FFLPYY192 pKa = 8.82MAKK195 pKa = 10.4LKK197 pKa = 11.05LSDD200 pKa = 3.67APIAYY205 pKa = 9.0RR206 pKa = 11.84AEE208 pKa = 4.08MATGGMSLINDD219 pKa = 4.21MCQSHH224 pKa = 7.36PGAKK228 pKa = 10.12YY229 pKa = 10.97LMTDD233 pKa = 3.74LSQFDD238 pKa = 3.79KK239 pKa = 11.16SVPPWLIRR247 pKa = 11.84DD248 pKa = 3.46AFGIVMEE255 pKa = 4.8NFDD258 pKa = 3.75FTRR261 pKa = 11.84VVGSDD266 pKa = 3.21GKK268 pKa = 10.97VWDD271 pKa = 4.15VNPDD275 pKa = 2.98RR276 pKa = 11.84TKK278 pKa = 10.91RR279 pKa = 11.84RR280 pKa = 11.84ISRR283 pKa = 11.84MIDD286 pKa = 3.12YY287 pKa = 9.08FINTPVRR294 pKa = 11.84LCNGEE299 pKa = 4.1RR300 pKa = 11.84YY301 pKa = 9.26RR302 pKa = 11.84KK303 pKa = 9.9RR304 pKa = 11.84GGVPSGSMWTNIVDD318 pKa = 4.49TIVNAIISRR327 pKa = 11.84YY328 pKa = 10.13CIFNTSGRR336 pKa = 11.84LPLADD341 pKa = 4.46MYY343 pKa = 11.55LGDD346 pKa = 4.22DD347 pKa = 3.36QFAVVDD353 pKa = 5.19GIINLNDD360 pKa = 3.02IAKK363 pKa = 10.4LMNEE367 pKa = 4.05SFGMVLHH374 pKa = 7.17PDD376 pKa = 2.87KK377 pKa = 11.07CVYY380 pKa = 9.44TEE382 pKa = 3.91NASNVQFLGYY392 pKa = 9.69FNRR395 pKa = 11.84NGLPWKK401 pKa = 10.88GNMFLVASMIFPEE414 pKa = 5.04RR415 pKa = 11.84PVNDD419 pKa = 2.61NMTRR423 pKa = 11.84VSRR426 pKa = 11.84AIGQMWSTLHH436 pKa = 6.64GGQAVRR442 pKa = 11.84WWNIVQAMLRR452 pKa = 11.84DD453 pKa = 4.0FGFTSKK459 pKa = 10.64EE460 pKa = 3.94VIEE463 pKa = 5.1NIQSRR468 pKa = 11.84PGSFRR473 pKa = 11.84YY474 pKa = 10.22LRR476 pKa = 11.84MLGIDD481 pKa = 3.18ISKK484 pKa = 10.22IGIPTRR490 pKa = 11.84YY491 pKa = 9.75GEE493 pKa = 4.63YY494 pKa = 10.0ILGIDD499 pKa = 4.66PPWKK503 pKa = 9.62SLKK506 pKa = 10.27SYY508 pKa = 10.95RR509 pKa = 11.84PVKK512 pKa = 9.97WDD514 pKa = 3.18IDD516 pKa = 3.8DD517 pKa = 4.28LDD519 pKa = 4.45RR520 pKa = 11.84RR521 pKa = 11.84SQSIDD526 pKa = 2.99MEE528 pKa = 4.13EE529 pKa = 3.46WHH531 pKa = 6.98NFIQCMDD538 pKa = 4.49DD539 pKa = 4.37SDD541 pKa = 5.99DD542 pKa = 3.73EE543 pKa = 4.4
MM1 pKa = 7.86EE2 pKa = 4.69YY3 pKa = 10.33LGKK6 pKa = 10.51AEE8 pKa = 4.47GFNMDD13 pKa = 4.36LRR15 pKa = 11.84PPRR18 pKa = 11.84MDD20 pKa = 5.92GITCGYY26 pKa = 8.48MNDD29 pKa = 3.84WFGDD33 pKa = 3.61AYY35 pKa = 11.26VDD37 pKa = 3.94VVMQTYY43 pKa = 9.99HH44 pKa = 7.24RR45 pKa = 11.84SQPSLEE51 pKa = 4.01LLKK54 pKa = 10.65EE55 pKa = 3.72DD56 pKa = 4.28FMRR59 pKa = 11.84YY60 pKa = 8.31DD61 pKa = 3.34RR62 pKa = 11.84KK63 pKa = 10.51YY64 pKa = 8.62VAGTVRR70 pKa = 11.84EE71 pKa = 4.08EE72 pKa = 3.48QSYY75 pKa = 11.03RR76 pKa = 11.84MVLQSVRR83 pKa = 11.84DD84 pKa = 3.67QFVLGEE90 pKa = 4.04KK91 pKa = 10.38LIPLTLGAVFEE102 pKa = 4.92SNSMTTDD109 pKa = 3.46KK110 pKa = 11.42SPGLPWIQRR119 pKa = 11.84GYY121 pKa = 8.11KK122 pKa = 9.04TKK124 pKa = 10.95GDD126 pKa = 3.35VFACHH131 pKa = 6.0EE132 pKa = 4.18ARR134 pKa = 11.84SEE136 pKa = 4.74IFRR139 pKa = 11.84TWDD142 pKa = 3.55RR143 pKa = 11.84IGNGRR148 pKa = 11.84GATLPDD154 pKa = 3.02TCAFMRR160 pKa = 11.84VQLAKK165 pKa = 10.3EE166 pKa = 4.22DD167 pKa = 3.75KK168 pKa = 10.22QKK170 pKa = 10.44IRR172 pKa = 11.84AVWGTPTDD180 pKa = 4.05VIAEE184 pKa = 4.22EE185 pKa = 4.29ARR187 pKa = 11.84FFLPYY192 pKa = 8.82MAKK195 pKa = 10.4LKK197 pKa = 11.05LSDD200 pKa = 3.67APIAYY205 pKa = 9.0RR206 pKa = 11.84AEE208 pKa = 4.08MATGGMSLINDD219 pKa = 4.21MCQSHH224 pKa = 7.36PGAKK228 pKa = 10.12YY229 pKa = 10.97LMTDD233 pKa = 3.74LSQFDD238 pKa = 3.79KK239 pKa = 11.16SVPPWLIRR247 pKa = 11.84DD248 pKa = 3.46AFGIVMEE255 pKa = 4.8NFDD258 pKa = 3.75FTRR261 pKa = 11.84VVGSDD266 pKa = 3.21GKK268 pKa = 10.97VWDD271 pKa = 4.15VNPDD275 pKa = 2.98RR276 pKa = 11.84TKK278 pKa = 10.91RR279 pKa = 11.84RR280 pKa = 11.84ISRR283 pKa = 11.84MIDD286 pKa = 3.12YY287 pKa = 9.08FINTPVRR294 pKa = 11.84LCNGEE299 pKa = 4.1RR300 pKa = 11.84YY301 pKa = 9.26RR302 pKa = 11.84KK303 pKa = 9.9RR304 pKa = 11.84GGVPSGSMWTNIVDD318 pKa = 4.49TIVNAIISRR327 pKa = 11.84YY328 pKa = 10.13CIFNTSGRR336 pKa = 11.84LPLADD341 pKa = 4.46MYY343 pKa = 11.55LGDD346 pKa = 4.22DD347 pKa = 3.36QFAVVDD353 pKa = 5.19GIINLNDD360 pKa = 3.02IAKK363 pKa = 10.4LMNEE367 pKa = 4.05SFGMVLHH374 pKa = 7.17PDD376 pKa = 2.87KK377 pKa = 11.07CVYY380 pKa = 9.44TEE382 pKa = 3.91NASNVQFLGYY392 pKa = 9.69FNRR395 pKa = 11.84NGLPWKK401 pKa = 10.88GNMFLVASMIFPEE414 pKa = 5.04RR415 pKa = 11.84PVNDD419 pKa = 2.61NMTRR423 pKa = 11.84VSRR426 pKa = 11.84AIGQMWSTLHH436 pKa = 6.64GGQAVRR442 pKa = 11.84WWNIVQAMLRR452 pKa = 11.84DD453 pKa = 4.0FGFTSKK459 pKa = 10.64EE460 pKa = 3.94VIEE463 pKa = 5.1NIQSRR468 pKa = 11.84PGSFRR473 pKa = 11.84YY474 pKa = 10.22LRR476 pKa = 11.84MLGIDD481 pKa = 3.18ISKK484 pKa = 10.22IGIPTRR490 pKa = 11.84YY491 pKa = 9.75GEE493 pKa = 4.63YY494 pKa = 10.0ILGIDD499 pKa = 4.66PPWKK503 pKa = 9.62SLKK506 pKa = 10.27SYY508 pKa = 10.95RR509 pKa = 11.84PVKK512 pKa = 9.97WDD514 pKa = 3.18IDD516 pKa = 3.8DD517 pKa = 4.28LDD519 pKa = 4.45RR520 pKa = 11.84RR521 pKa = 11.84SQSIDD526 pKa = 2.99MEE528 pKa = 4.13EE529 pKa = 3.46WHH531 pKa = 6.98NFIQCMDD538 pKa = 4.49DD539 pKa = 4.37SDD541 pKa = 5.99DD542 pKa = 3.73EE543 pKa = 4.4
Molecular weight: 62.4 kDa
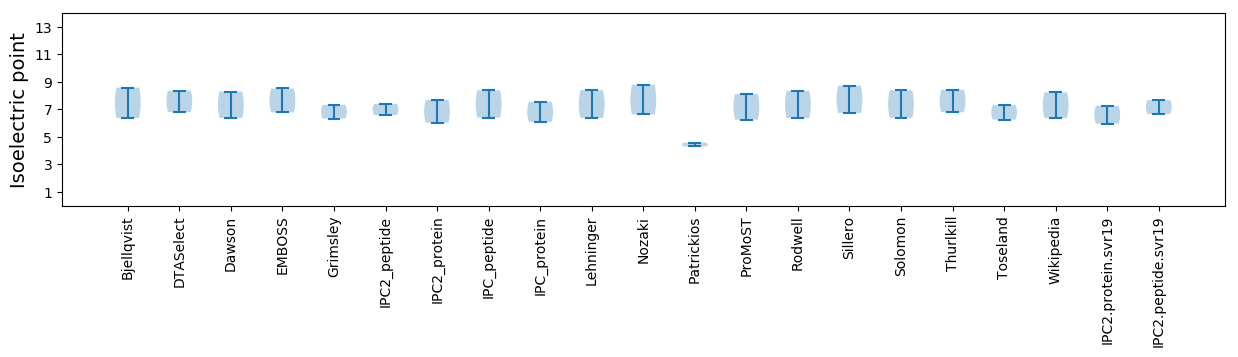
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLP6|A0A1L3KLP6_9VIRU RdRp OS=Hubei partiti-like virus 11 OX=1923017 PE=4 SV=1
MM1 pKa = 6.84MSEE4 pKa = 4.29RR5 pKa = 11.84SHH7 pKa = 7.23LNLKK11 pKa = 9.64MNTDD15 pKa = 3.75KK16 pKa = 10.4KK17 pKa = 9.18TKK19 pKa = 10.41SKK21 pKa = 10.91GPKK24 pKa = 8.39KK25 pKa = 8.56TSKK28 pKa = 10.33PKK30 pKa = 9.97EE31 pKa = 4.15EE32 pKa = 4.15KK33 pKa = 10.27KK34 pKa = 10.83EE35 pKa = 3.97PVDD38 pKa = 3.73APSVKK43 pKa = 10.41VEE45 pKa = 3.95VEE47 pKa = 3.95KK48 pKa = 10.81PKK50 pKa = 10.05PAEE53 pKa = 3.87VTKK56 pKa = 9.92PAEE59 pKa = 4.18PVKK62 pKa = 9.37PTPAPQKK69 pKa = 10.21SKK71 pKa = 10.88PKK73 pKa = 9.97AQSQVASKK81 pKa = 10.27LQNDD85 pKa = 4.58PLWEE89 pKa = 4.57DD90 pKa = 3.89FGSDD94 pKa = 3.91DD95 pKa = 5.06FDD97 pKa = 3.93VLMPEE102 pKa = 4.19RR103 pKa = 11.84GDD105 pKa = 3.2NRR107 pKa = 11.84FAVTLEE113 pKa = 4.15GYY115 pKa = 9.98IPLIEE120 pKa = 4.1QTYY123 pKa = 7.64EE124 pKa = 4.1TLSHH128 pKa = 6.56EE129 pKa = 4.86DD130 pKa = 3.16KK131 pKa = 11.07TFGKK135 pKa = 10.58SMSLSMFSWYY145 pKa = 10.02CCQHH149 pKa = 7.34LYY151 pKa = 9.81MRR153 pKa = 11.84IITIRR158 pKa = 11.84MSEE161 pKa = 4.09GLATSDD167 pKa = 3.16EE168 pKa = 4.07KK169 pKa = 11.21RR170 pKa = 11.84YY171 pKa = 9.14VAEE174 pKa = 3.98VSSIPFVIPLTIEE187 pKa = 4.07IYY189 pKa = 10.57LKK191 pKa = 10.9SIGNVMDD198 pKa = 4.91AEE200 pKa = 4.37LNKK203 pKa = 10.78LKK205 pKa = 10.44ISFPAFPNAEE215 pKa = 3.84GHH217 pKa = 6.62FGVVNNNSHH226 pKa = 5.89WKK228 pKa = 9.17YY229 pKa = 10.08EE230 pKa = 4.44SVPAPYY236 pKa = 8.71VTSRR240 pKa = 11.84RR241 pKa = 11.84MIHH244 pKa = 7.12DD245 pKa = 3.58YY246 pKa = 10.97AYY248 pKa = 7.37TTGLEE253 pKa = 4.0NDD255 pKa = 5.04RR256 pKa = 11.84IWTLPLNLLPPNRR269 pKa = 11.84DD270 pKa = 3.0ATRR273 pKa = 11.84HH274 pKa = 6.06RR275 pKa = 11.84ICGNPTKK282 pKa = 10.81NLLGWARR289 pKa = 11.84SSMLTSEE296 pKa = 3.87QTLILEE302 pKa = 4.39SCGIDD307 pKa = 3.3GANFPTEE314 pKa = 3.46RR315 pKa = 11.84PMFQFNRR322 pKa = 11.84GLFVLLSSHH331 pKa = 6.87LSAASRR337 pKa = 11.84SMKK340 pKa = 9.88MSGSAITSIEE350 pKa = 3.92GSFGQTLFVEE360 pKa = 4.67RR361 pKa = 11.84DD362 pKa = 3.28DD363 pKa = 3.94RR364 pKa = 11.84HH365 pKa = 6.58PEE367 pKa = 3.7EE368 pKa = 4.46FDD370 pKa = 4.87RR371 pKa = 11.84NVQYY375 pKa = 11.36CEE377 pKa = 3.88MGNLCTVSAFQVDD390 pKa = 3.36KK391 pKa = 11.06RR392 pKa = 11.84FSQGAAVCNFRR403 pKa = 11.84SRR405 pKa = 11.84KK406 pKa = 9.6FEE408 pKa = 4.22INNLRR413 pKa = 11.84SYY415 pKa = 11.6ACFDD419 pKa = 3.71FGDD422 pKa = 4.3YY423 pKa = 10.96EE424 pKa = 4.28HH425 pKa = 7.73VPDD428 pKa = 4.06GWNNTVNTIFDD439 pKa = 4.36FGAQADD445 pKa = 3.77WNRR448 pKa = 11.84PKK450 pKa = 9.86FTSAYY455 pKa = 7.93GSKK458 pKa = 10.31VALRR462 pKa = 11.84QSWVKK467 pKa = 10.56KK468 pKa = 10.24SLVRR472 pKa = 11.84DD473 pKa = 3.56KK474 pKa = 10.99TT475 pKa = 3.62
MM1 pKa = 6.84MSEE4 pKa = 4.29RR5 pKa = 11.84SHH7 pKa = 7.23LNLKK11 pKa = 9.64MNTDD15 pKa = 3.75KK16 pKa = 10.4KK17 pKa = 9.18TKK19 pKa = 10.41SKK21 pKa = 10.91GPKK24 pKa = 8.39KK25 pKa = 8.56TSKK28 pKa = 10.33PKK30 pKa = 9.97EE31 pKa = 4.15EE32 pKa = 4.15KK33 pKa = 10.27KK34 pKa = 10.83EE35 pKa = 3.97PVDD38 pKa = 3.73APSVKK43 pKa = 10.41VEE45 pKa = 3.95VEE47 pKa = 3.95KK48 pKa = 10.81PKK50 pKa = 10.05PAEE53 pKa = 3.87VTKK56 pKa = 9.92PAEE59 pKa = 4.18PVKK62 pKa = 9.37PTPAPQKK69 pKa = 10.21SKK71 pKa = 10.88PKK73 pKa = 9.97AQSQVASKK81 pKa = 10.27LQNDD85 pKa = 4.58PLWEE89 pKa = 4.57DD90 pKa = 3.89FGSDD94 pKa = 3.91DD95 pKa = 5.06FDD97 pKa = 3.93VLMPEE102 pKa = 4.19RR103 pKa = 11.84GDD105 pKa = 3.2NRR107 pKa = 11.84FAVTLEE113 pKa = 4.15GYY115 pKa = 9.98IPLIEE120 pKa = 4.1QTYY123 pKa = 7.64EE124 pKa = 4.1TLSHH128 pKa = 6.56EE129 pKa = 4.86DD130 pKa = 3.16KK131 pKa = 11.07TFGKK135 pKa = 10.58SMSLSMFSWYY145 pKa = 10.02CCQHH149 pKa = 7.34LYY151 pKa = 9.81MRR153 pKa = 11.84IITIRR158 pKa = 11.84MSEE161 pKa = 4.09GLATSDD167 pKa = 3.16EE168 pKa = 4.07KK169 pKa = 11.21RR170 pKa = 11.84YY171 pKa = 9.14VAEE174 pKa = 3.98VSSIPFVIPLTIEE187 pKa = 4.07IYY189 pKa = 10.57LKK191 pKa = 10.9SIGNVMDD198 pKa = 4.91AEE200 pKa = 4.37LNKK203 pKa = 10.78LKK205 pKa = 10.44ISFPAFPNAEE215 pKa = 3.84GHH217 pKa = 6.62FGVVNNNSHH226 pKa = 5.89WKK228 pKa = 9.17YY229 pKa = 10.08EE230 pKa = 4.44SVPAPYY236 pKa = 8.71VTSRR240 pKa = 11.84RR241 pKa = 11.84MIHH244 pKa = 7.12DD245 pKa = 3.58YY246 pKa = 10.97AYY248 pKa = 7.37TTGLEE253 pKa = 4.0NDD255 pKa = 5.04RR256 pKa = 11.84IWTLPLNLLPPNRR269 pKa = 11.84DD270 pKa = 3.0ATRR273 pKa = 11.84HH274 pKa = 6.06RR275 pKa = 11.84ICGNPTKK282 pKa = 10.81NLLGWARR289 pKa = 11.84SSMLTSEE296 pKa = 3.87QTLILEE302 pKa = 4.39SCGIDD307 pKa = 3.3GANFPTEE314 pKa = 3.46RR315 pKa = 11.84PMFQFNRR322 pKa = 11.84GLFVLLSSHH331 pKa = 6.87LSAASRR337 pKa = 11.84SMKK340 pKa = 9.88MSGSAITSIEE350 pKa = 3.92GSFGQTLFVEE360 pKa = 4.67RR361 pKa = 11.84DD362 pKa = 3.28DD363 pKa = 3.94RR364 pKa = 11.84HH365 pKa = 6.58PEE367 pKa = 3.7EE368 pKa = 4.46FDD370 pKa = 4.87RR371 pKa = 11.84NVQYY375 pKa = 11.36CEE377 pKa = 3.88MGNLCTVSAFQVDD390 pKa = 3.36KK391 pKa = 11.06RR392 pKa = 11.84FSQGAAVCNFRR403 pKa = 11.84SRR405 pKa = 11.84KK406 pKa = 9.6FEE408 pKa = 4.22INNLRR413 pKa = 11.84SYY415 pKa = 11.6ACFDD419 pKa = 3.71FGDD422 pKa = 4.3YY423 pKa = 10.96EE424 pKa = 4.28HH425 pKa = 7.73VPDD428 pKa = 4.06GWNNTVNTIFDD439 pKa = 4.36FGAQADD445 pKa = 3.77WNRR448 pKa = 11.84PKK450 pKa = 9.86FTSAYY455 pKa = 7.93GSKK458 pKa = 10.31VALRR462 pKa = 11.84QSWVKK467 pKa = 10.56KK468 pKa = 10.24SLVRR472 pKa = 11.84DD473 pKa = 3.56KK474 pKa = 10.99TT475 pKa = 3.62
Molecular weight: 53.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1018 |
475 |
543 |
509.0 |
58.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.501 ± 0.256 | 1.572 ± 0.073 |
6.778 ± 0.986 | 5.599 ± 0.74 |
5.01 ± 0.165 | 6.385 ± 0.867 |
1.572 ± 0.347 | 5.501 ± 0.84 |
5.992 ± 1.032 | 6.974 ± 0.119 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.224 ± 0.694 | 5.01 ± 0.302 |
5.501 ± 0.53 | 3.143 ± 0.128 |
6.68 ± 0.785 | 7.564 ± 1.106 |
5.206 ± 0.448 | 6.189 ± 0.191 |
2.161 ± 0.31 | 3.438 ± 0.319 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
