
Puccinia sorghi
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Pucciniomycotina; Pucciniomycetes; Pucciniales; Pucciniaceae; Puccinia
Average proteome isoelectric point is 7.51
Get precalculated fractions of proteins
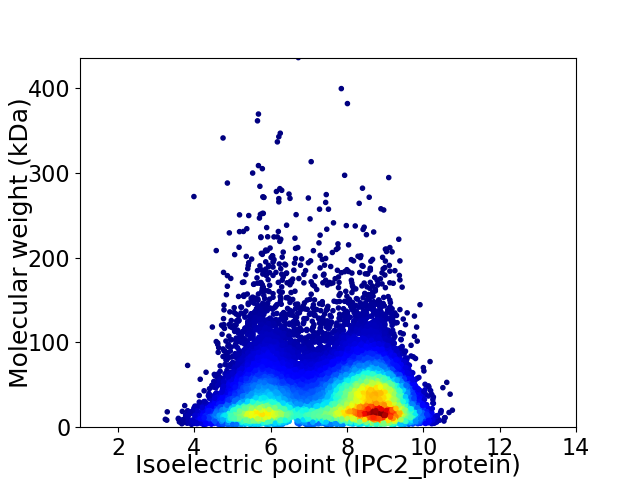
Virtual 2D-PAGE plot for 21032 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>tr|A0A0L6UEM3|A0A0L6UEM3_9BASI Uncharacterized protein OS=Puccinia sorghi OX=27349 GN=VP01_679g1 PE=4 SV=1
CC1 pKa = 7.31FFDD4 pKa = 5.79KK5 pKa = 11.23LIDD8 pKa = 5.4LDD10 pKa = 4.29DD11 pKa = 3.53TDD13 pKa = 3.59YY14 pKa = 11.44WFDD17 pKa = 4.46FDD19 pKa = 5.23VLGCAEE25 pKa = 4.19TRR27 pKa = 11.84NLDD30 pKa = 3.87SIADD34 pKa = 4.19LSCPTNPSPADD45 pKa = 3.89RR46 pKa = 11.84DD47 pKa = 3.82PLLVEE52 pKa = 3.79QDD54 pKa = 3.73PLRR57 pKa = 11.84EE58 pKa = 3.92ARR60 pKa = 11.84QLDD63 pKa = 3.79EE64 pKa = 5.33GVCTSNDD71 pKa = 4.02PIGCSVSEE79 pKa = 4.41TNSTLPAPQYY89 pKa = 11.09CEE91 pKa = 3.7FVLYY95 pKa = 10.95SLVPVSAGQPSPPCEE110 pKa = 3.37IWKK113 pKa = 9.88DD114 pKa = 3.57AEE116 pKa = 4.34IVDD119 pKa = 4.62TICRR123 pKa = 11.84EE124 pKa = 4.17GPADD128 pKa = 3.7RR129 pKa = 11.84TTPRR133 pKa = 3.53
CC1 pKa = 7.31FFDD4 pKa = 5.79KK5 pKa = 11.23LIDD8 pKa = 5.4LDD10 pKa = 4.29DD11 pKa = 3.53TDD13 pKa = 3.59YY14 pKa = 11.44WFDD17 pKa = 4.46FDD19 pKa = 5.23VLGCAEE25 pKa = 4.19TRR27 pKa = 11.84NLDD30 pKa = 3.87SIADD34 pKa = 4.19LSCPTNPSPADD45 pKa = 3.89RR46 pKa = 11.84DD47 pKa = 3.82PLLVEE52 pKa = 3.79QDD54 pKa = 3.73PLRR57 pKa = 11.84EE58 pKa = 3.92ARR60 pKa = 11.84QLDD63 pKa = 3.79EE64 pKa = 5.33GVCTSNDD71 pKa = 4.02PIGCSVSEE79 pKa = 4.41TNSTLPAPQYY89 pKa = 11.09CEE91 pKa = 3.7FVLYY95 pKa = 10.95SLVPVSAGQPSPPCEE110 pKa = 3.37IWKK113 pKa = 9.88DD114 pKa = 3.57AEE116 pKa = 4.34IVDD119 pKa = 4.62TICRR123 pKa = 11.84EE124 pKa = 4.17GPADD128 pKa = 3.7RR129 pKa = 11.84TTPRR133 pKa = 3.53
Molecular weight: 14.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0L6V4K4|A0A0L6V4K4_9BASI Uncharacterized protein OS=Puccinia sorghi OX=27349 GN=VP01_2682g3 PE=4 SV=1
MM1 pKa = 8.12RR2 pKa = 11.84PRR4 pKa = 11.84KK5 pKa = 9.81KK6 pKa = 10.67NGVFLIAFTSGQFIIVAARR25 pKa = 11.84RR26 pKa = 11.84LPRR29 pKa = 11.84RR30 pKa = 11.84EE31 pKa = 3.88QKK33 pKa = 10.43HH34 pKa = 5.18KK35 pKa = 10.55SRR37 pKa = 11.84TNRR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 4.96AGEE45 pKa = 4.17RR46 pKa = 11.84KK47 pKa = 9.59LRR49 pKa = 11.84TAWLGEE55 pKa = 3.67GRR57 pKa = 11.84GLLGEE62 pKa = 4.2THH64 pKa = 5.6EE65 pKa = 4.35QEE67 pKa = 4.49HH68 pKa = 6.08LHH70 pKa = 7.06AEE72 pKa = 4.12HH73 pKa = 7.33ADD75 pKa = 3.67NGGEE79 pKa = 3.66EE80 pKa = 4.08HH81 pKa = 7.18FYY83 pKa = 10.66IKK85 pKa = 10.73YY86 pKa = 10.0ILIVVLKK93 pKa = 10.36RR94 pKa = 11.84LVRR97 pKa = 11.84LKK99 pKa = 10.38RR100 pKa = 11.84PMMMEE105 pKa = 4.2SSSHH109 pKa = 5.22GACRR113 pKa = 11.84RR114 pKa = 11.84CCWPIGGKK122 pKa = 10.42AMGSGPTYY130 pKa = 8.12RR131 pKa = 11.84TRR133 pKa = 11.84RR134 pKa = 11.84GSEE137 pKa = 3.75LATCRR142 pKa = 11.84QSDD145 pKa = 4.05PSVKK149 pKa = 9.79TPSQSGLLTCRR160 pKa = 11.84GQCCGLRR167 pKa = 11.84TACHH171 pKa = 6.33SSLSVGGDD179 pKa = 3.4YY180 pKa = 10.92LSPPGHH186 pKa = 6.57SYY188 pKa = 11.41GILNRR193 pKa = 11.84EE194 pKa = 4.63LFPQQLGNTIGTATYY209 pKa = 9.5FDD211 pKa = 4.34SRR213 pKa = 11.84AEE215 pKa = 4.14SPLNALSIGQDD226 pKa = 2.78ASGVVTRR233 pKa = 11.84CLTPQICLDD242 pKa = 3.73VNQKK246 pKa = 8.64STVRR250 pKa = 11.84QLLSNSIRR258 pKa = 11.84PHH260 pKa = 6.38PGRR263 pKa = 11.84GVGSTASGQGKK274 pKa = 7.54WPSGPVVVRR283 pKa = 11.84FDD285 pKa = 3.4IVKK288 pKa = 10.39SVANQATTFDD298 pKa = 3.86VSPPASVIKK307 pKa = 10.31GGRR310 pKa = 11.84PLTVMLISFSLTTRR324 pKa = 11.84PLRR327 pKa = 11.84RR328 pKa = 11.84CFSKK332 pKa = 10.72RR333 pKa = 11.84GFNSSKK339 pKa = 10.43MMLEE343 pKa = 4.56QQDD346 pKa = 3.84SWCGRR351 pKa = 11.84PSHH354 pKa = 6.21QPAPMVSYY362 pKa = 10.06PLVSVGLTAKK372 pKa = 9.22LQKK375 pKa = 9.29MRR377 pKa = 11.84CCAPGSEE384 pKa = 4.04VRR386 pKa = 11.84LRR388 pKa = 11.84AGRR391 pKa = 11.84VTQHH395 pKa = 5.79VSMHH399 pKa = 6.45TILRR403 pKa = 11.84CC404 pKa = 3.6
MM1 pKa = 8.12RR2 pKa = 11.84PRR4 pKa = 11.84KK5 pKa = 9.81KK6 pKa = 10.67NGVFLIAFTSGQFIIVAARR25 pKa = 11.84RR26 pKa = 11.84LPRR29 pKa = 11.84RR30 pKa = 11.84EE31 pKa = 3.88QKK33 pKa = 10.43HH34 pKa = 5.18KK35 pKa = 10.55SRR37 pKa = 11.84TNRR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 4.96AGEE45 pKa = 4.17RR46 pKa = 11.84KK47 pKa = 9.59LRR49 pKa = 11.84TAWLGEE55 pKa = 3.67GRR57 pKa = 11.84GLLGEE62 pKa = 4.2THH64 pKa = 5.6EE65 pKa = 4.35QEE67 pKa = 4.49HH68 pKa = 6.08LHH70 pKa = 7.06AEE72 pKa = 4.12HH73 pKa = 7.33ADD75 pKa = 3.67NGGEE79 pKa = 3.66EE80 pKa = 4.08HH81 pKa = 7.18FYY83 pKa = 10.66IKK85 pKa = 10.73YY86 pKa = 10.0ILIVVLKK93 pKa = 10.36RR94 pKa = 11.84LVRR97 pKa = 11.84LKK99 pKa = 10.38RR100 pKa = 11.84PMMMEE105 pKa = 4.2SSSHH109 pKa = 5.22GACRR113 pKa = 11.84RR114 pKa = 11.84CCWPIGGKK122 pKa = 10.42AMGSGPTYY130 pKa = 8.12RR131 pKa = 11.84TRR133 pKa = 11.84RR134 pKa = 11.84GSEE137 pKa = 3.75LATCRR142 pKa = 11.84QSDD145 pKa = 4.05PSVKK149 pKa = 9.79TPSQSGLLTCRR160 pKa = 11.84GQCCGLRR167 pKa = 11.84TACHH171 pKa = 6.33SSLSVGGDD179 pKa = 3.4YY180 pKa = 10.92LSPPGHH186 pKa = 6.57SYY188 pKa = 11.41GILNRR193 pKa = 11.84EE194 pKa = 4.63LFPQQLGNTIGTATYY209 pKa = 9.5FDD211 pKa = 4.34SRR213 pKa = 11.84AEE215 pKa = 4.14SPLNALSIGQDD226 pKa = 2.78ASGVVTRR233 pKa = 11.84CLTPQICLDD242 pKa = 3.73VNQKK246 pKa = 8.64STVRR250 pKa = 11.84QLLSNSIRR258 pKa = 11.84PHH260 pKa = 6.38PGRR263 pKa = 11.84GVGSTASGQGKK274 pKa = 7.54WPSGPVVVRR283 pKa = 11.84FDD285 pKa = 3.4IVKK288 pKa = 10.39SVANQATTFDD298 pKa = 3.86VSPPASVIKK307 pKa = 10.31GGRR310 pKa = 11.84PLTVMLISFSLTTRR324 pKa = 11.84PLRR327 pKa = 11.84RR328 pKa = 11.84CFSKK332 pKa = 10.72RR333 pKa = 11.84GFNSSKK339 pKa = 10.43MMLEE343 pKa = 4.56QQDD346 pKa = 3.84SWCGRR351 pKa = 11.84PSHH354 pKa = 6.21QPAPMVSYY362 pKa = 10.06PLVSVGLTAKK372 pKa = 9.22LQKK375 pKa = 9.29MRR377 pKa = 11.84CCAPGSEE384 pKa = 4.04VRR386 pKa = 11.84LRR388 pKa = 11.84AGRR391 pKa = 11.84VTQHH395 pKa = 5.79VSMHH399 pKa = 6.45TILRR403 pKa = 11.84CC404 pKa = 3.6
Molecular weight: 44.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7432697 |
28 |
3846 |
353.4 |
39.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.112 ± 0.019 | 2.065 ± 0.011 |
4.348 ± 0.013 | 5.008 ± 0.016 |
4.611 ± 0.016 | 5.309 ± 0.016 |
3.243 ± 0.011 | 5.74 ± 0.014 |
5.897 ± 0.019 | 10.367 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.114 ± 0.006 | 4.654 ± 0.011 |
6.179 ± 0.022 | 4.593 ± 0.012 |
5.165 ± 0.014 | 9.686 ± 0.023 |
5.794 ± 0.01 | 5.105 ± 0.011 |
1.433 ± 0.007 | 2.574 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
