
Ambe virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Ambe phlebovirus
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
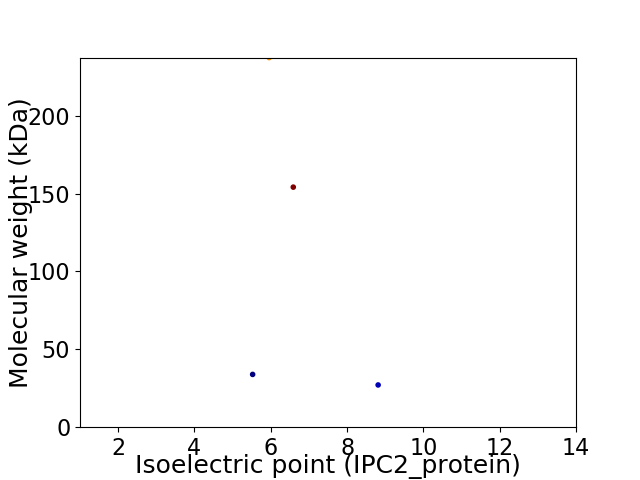
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1S5SHU8|A0A1S5SHU8_9VIRU Nucleoprotein OS=Ambe virus OX=1926500 PE=3 SV=1
MM1 pKa = 7.41NKK3 pKa = 9.84FYY5 pKa = 10.6LYY7 pKa = 10.96DD8 pKa = 3.49KK9 pKa = 10.38MSIIRR14 pKa = 11.84PGGLYY19 pKa = 10.45CSLPRR24 pKa = 11.84VTFEE28 pKa = 5.08AFNAYY33 pKa = 9.1QEE35 pKa = 4.48NPISRR40 pKa = 11.84YY41 pKa = 9.75SDD43 pKa = 3.02IEE45 pKa = 4.12FPLPAGFRR53 pKa = 11.84VSSKK57 pKa = 9.09TRR59 pKa = 11.84ATLSDD64 pKa = 3.71YY65 pKa = 11.43YY66 pKa = 10.35MNRR69 pKa = 11.84EE70 pKa = 3.86FPMHH74 pKa = 6.79WGCSQQSYY82 pKa = 11.07ALAPYY87 pKa = 8.5ITVFDD92 pKa = 4.2EE93 pKa = 4.67MIEE96 pKa = 4.22CLSHH100 pKa = 6.84FKK102 pKa = 10.27EE103 pKa = 4.42HH104 pKa = 6.82EE105 pKa = 4.0ISAEE109 pKa = 3.94CHH111 pKa = 6.07PNLHH115 pKa = 6.66EE116 pKa = 4.47ALSWPTGGPNLDD128 pKa = 3.33YY129 pKa = 10.87MRR131 pKa = 11.84FYY133 pKa = 11.28SNKK136 pKa = 8.08PDD138 pKa = 3.14CMYY141 pKa = 10.73EE142 pKa = 4.2RR143 pKa = 11.84KK144 pKa = 9.84CRR146 pKa = 11.84EE147 pKa = 3.46ATLILRR153 pKa = 11.84ASRR156 pKa = 11.84PNWWFAEE163 pKa = 4.02SLYY166 pKa = 10.78NSHH169 pKa = 6.93KK170 pKa = 10.38NAFHH174 pKa = 6.49EE175 pKa = 4.7SVVRR179 pKa = 11.84GLDD182 pKa = 3.15PRR184 pKa = 11.84FFNGSDD190 pKa = 3.63VIKK193 pKa = 9.67EE194 pKa = 3.8ICIVQCVRR202 pKa = 11.84LLNAVLFDD210 pKa = 4.65GIYY213 pKa = 10.61SRR215 pKa = 11.84VPNKK219 pKa = 9.96FYY221 pKa = 10.42MAVEE225 pKa = 3.79YY226 pKa = 10.46FRR228 pKa = 11.84NKK230 pKa = 10.34LEE232 pKa = 4.18HH233 pKa = 6.95LGPAILGNRR242 pKa = 11.84KK243 pKa = 8.98WIPHH247 pKa = 4.66VNTRR251 pKa = 11.84FDD253 pKa = 4.62LLNKK257 pKa = 10.39CIDD260 pKa = 3.73EE261 pKa = 4.33EE262 pKa = 4.63FCTDD266 pKa = 3.28SVYY269 pKa = 10.98EE270 pKa = 4.17SDD272 pKa = 5.16LDD274 pKa = 3.92LAKK277 pKa = 10.67EE278 pKa = 4.16FEE280 pKa = 5.97DD281 pKa = 3.24ILKK284 pKa = 10.68DD285 pKa = 3.62VTEE288 pKa = 4.3LL289 pKa = 3.64
MM1 pKa = 7.41NKK3 pKa = 9.84FYY5 pKa = 10.6LYY7 pKa = 10.96DD8 pKa = 3.49KK9 pKa = 10.38MSIIRR14 pKa = 11.84PGGLYY19 pKa = 10.45CSLPRR24 pKa = 11.84VTFEE28 pKa = 5.08AFNAYY33 pKa = 9.1QEE35 pKa = 4.48NPISRR40 pKa = 11.84YY41 pKa = 9.75SDD43 pKa = 3.02IEE45 pKa = 4.12FPLPAGFRR53 pKa = 11.84VSSKK57 pKa = 9.09TRR59 pKa = 11.84ATLSDD64 pKa = 3.71YY65 pKa = 11.43YY66 pKa = 10.35MNRR69 pKa = 11.84EE70 pKa = 3.86FPMHH74 pKa = 6.79WGCSQQSYY82 pKa = 11.07ALAPYY87 pKa = 8.5ITVFDD92 pKa = 4.2EE93 pKa = 4.67MIEE96 pKa = 4.22CLSHH100 pKa = 6.84FKK102 pKa = 10.27EE103 pKa = 4.42HH104 pKa = 6.82EE105 pKa = 4.0ISAEE109 pKa = 3.94CHH111 pKa = 6.07PNLHH115 pKa = 6.66EE116 pKa = 4.47ALSWPTGGPNLDD128 pKa = 3.33YY129 pKa = 10.87MRR131 pKa = 11.84FYY133 pKa = 11.28SNKK136 pKa = 8.08PDD138 pKa = 3.14CMYY141 pKa = 10.73EE142 pKa = 4.2RR143 pKa = 11.84KK144 pKa = 9.84CRR146 pKa = 11.84EE147 pKa = 3.46ATLILRR153 pKa = 11.84ASRR156 pKa = 11.84PNWWFAEE163 pKa = 4.02SLYY166 pKa = 10.78NSHH169 pKa = 6.93KK170 pKa = 10.38NAFHH174 pKa = 6.49EE175 pKa = 4.7SVVRR179 pKa = 11.84GLDD182 pKa = 3.15PRR184 pKa = 11.84FFNGSDD190 pKa = 3.63VIKK193 pKa = 9.67EE194 pKa = 3.8ICIVQCVRR202 pKa = 11.84LLNAVLFDD210 pKa = 4.65GIYY213 pKa = 10.61SRR215 pKa = 11.84VPNKK219 pKa = 9.96FYY221 pKa = 10.42MAVEE225 pKa = 3.79YY226 pKa = 10.46FRR228 pKa = 11.84NKK230 pKa = 10.34LEE232 pKa = 4.18HH233 pKa = 6.95LGPAILGNRR242 pKa = 11.84KK243 pKa = 8.98WIPHH247 pKa = 4.66VNTRR251 pKa = 11.84FDD253 pKa = 4.62LLNKK257 pKa = 10.39CIDD260 pKa = 3.73EE261 pKa = 4.33EE262 pKa = 4.63FCTDD266 pKa = 3.28SVYY269 pKa = 10.98EE270 pKa = 4.17SDD272 pKa = 5.16LDD274 pKa = 3.92LAKK277 pKa = 10.67EE278 pKa = 4.16FEE280 pKa = 5.97DD281 pKa = 3.24ILKK284 pKa = 10.68DD285 pKa = 3.62VTEE288 pKa = 4.3LL289 pKa = 3.64
Molecular weight: 33.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S5SHU8|A0A1S5SHU8_9VIRU Nucleoprotein OS=Ambe virus OX=1926500 PE=3 SV=1
MM1 pKa = 7.8TDD3 pKa = 3.44YY4 pKa = 10.82QALAIEE10 pKa = 4.66FANAGVSPAEE20 pKa = 3.68IQDD23 pKa = 3.46WVRR26 pKa = 11.84EE27 pKa = 3.97FAYY30 pKa = 10.46QGFDD34 pKa = 2.66AHH36 pKa = 7.34RR37 pKa = 11.84IIEE40 pKa = 4.16LLRR43 pKa = 11.84TLGGDD48 pKa = 2.96AWQADD53 pKa = 4.1ARR55 pKa = 11.84RR56 pKa = 11.84MIVLGLTRR64 pKa = 11.84GNKK67 pKa = 8.66LEE69 pKa = 4.02KK70 pKa = 9.67MVMRR74 pKa = 11.84MSDD77 pKa = 2.95KK78 pKa = 10.92GKK80 pKa = 9.86QATADD85 pKa = 3.64LVRR88 pKa = 11.84KK89 pKa = 9.86YY90 pKa = 10.37KK91 pKa = 10.65LKK93 pKa = 10.72SGNPSRR99 pKa = 11.84DD100 pKa = 3.28EE101 pKa = 3.99LTLSRR106 pKa = 11.84VAAALAGWTCQALVAVQEE124 pKa = 4.33HH125 pKa = 6.26LPITGSYY132 pKa = 8.43MDD134 pKa = 4.61SVSPGYY140 pKa = 8.75PRR142 pKa = 11.84QMMHH146 pKa = 7.0PSFAGLVDD154 pKa = 3.21PTIPQTHH161 pKa = 7.46LDD163 pKa = 3.91TVLEE167 pKa = 4.03AHH169 pKa = 6.47SLYY172 pKa = 10.56LVHH175 pKa = 7.3FSRR178 pKa = 11.84MINPEE183 pKa = 3.45LRR185 pKa = 11.84GRR187 pKa = 11.84PRR189 pKa = 11.84SEE191 pKa = 3.78VVATFRR197 pKa = 11.84QPMLAAVNSNFITPEE212 pKa = 3.51QRR214 pKa = 11.84RR215 pKa = 11.84KK216 pKa = 9.81FLVSFGIVNSNGVPSAQVTAAANVFKK242 pKa = 11.13AMPP245 pKa = 4.23
MM1 pKa = 7.8TDD3 pKa = 3.44YY4 pKa = 10.82QALAIEE10 pKa = 4.66FANAGVSPAEE20 pKa = 3.68IQDD23 pKa = 3.46WVRR26 pKa = 11.84EE27 pKa = 3.97FAYY30 pKa = 10.46QGFDD34 pKa = 2.66AHH36 pKa = 7.34RR37 pKa = 11.84IIEE40 pKa = 4.16LLRR43 pKa = 11.84TLGGDD48 pKa = 2.96AWQADD53 pKa = 4.1ARR55 pKa = 11.84RR56 pKa = 11.84MIVLGLTRR64 pKa = 11.84GNKK67 pKa = 8.66LEE69 pKa = 4.02KK70 pKa = 9.67MVMRR74 pKa = 11.84MSDD77 pKa = 2.95KK78 pKa = 10.92GKK80 pKa = 9.86QATADD85 pKa = 3.64LVRR88 pKa = 11.84KK89 pKa = 9.86YY90 pKa = 10.37KK91 pKa = 10.65LKK93 pKa = 10.72SGNPSRR99 pKa = 11.84DD100 pKa = 3.28EE101 pKa = 3.99LTLSRR106 pKa = 11.84VAAALAGWTCQALVAVQEE124 pKa = 4.33HH125 pKa = 6.26LPITGSYY132 pKa = 8.43MDD134 pKa = 4.61SVSPGYY140 pKa = 8.75PRR142 pKa = 11.84QMMHH146 pKa = 7.0PSFAGLVDD154 pKa = 3.21PTIPQTHH161 pKa = 7.46LDD163 pKa = 3.91TVLEE167 pKa = 4.03AHH169 pKa = 6.47SLYY172 pKa = 10.56LVHH175 pKa = 7.3FSRR178 pKa = 11.84MINPEE183 pKa = 3.45LRR185 pKa = 11.84GRR187 pKa = 11.84PRR189 pKa = 11.84SEE191 pKa = 3.78VVATFRR197 pKa = 11.84QPMLAAVNSNFITPEE212 pKa = 3.51QRR214 pKa = 11.84RR215 pKa = 11.84KK216 pKa = 9.81FLVSFGIVNSNGVPSAQVTAAANVFKK242 pKa = 11.13AMPP245 pKa = 4.23
Molecular weight: 27.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3997 |
245 |
2087 |
999.3 |
113.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.154 ± 1.02 | 2.602 ± 0.721 |
5.154 ± 0.65 | 7.105 ± 0.438 |
5.279 ± 0.313 | 5.554 ± 0.248 |
2.627 ± 0.075 | 6.48 ± 0.476 |
6.805 ± 0.712 | 9.057 ± 0.099 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.177 ± 0.494 | 4.528 ± 0.361 |
3.728 ± 0.529 | 3.227 ± 0.306 |
4.929 ± 0.537 | 8.982 ± 0.693 |
5.504 ± 0.566 | 6.28 ± 0.285 |
1.176 ± 0.154 | 2.652 ± 0.518 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
