
Vicugna pacos polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
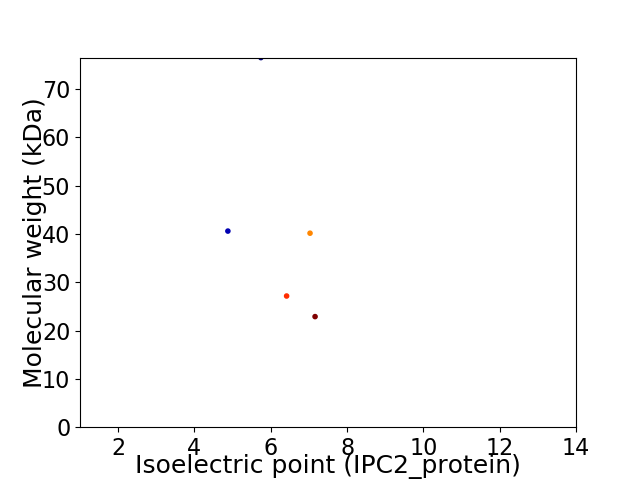
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
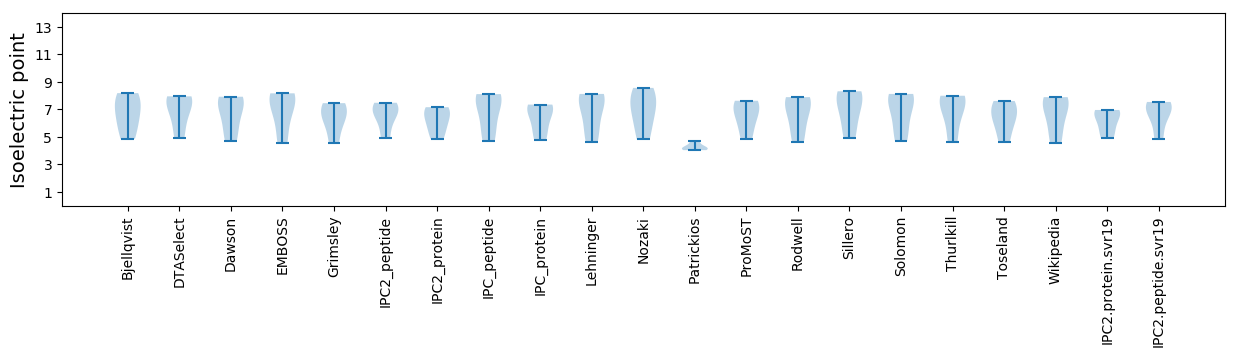
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V0CLV5|A0A1V0CLV5_9POLY Small t antigen OS=Vicugna pacos polyomavirus 1 OX=2169757 PE=4 SV=1
MM1 pKa = 7.41GALLAIPEE9 pKa = 4.53IIAAFSIAGAEE20 pKa = 4.02ALEE23 pKa = 4.17ITGGSLAAIAAGEE36 pKa = 4.06GLAAAEE42 pKa = 4.51ALTEE46 pKa = 4.13SAALIGSEE54 pKa = 4.21ALTMSEE60 pKa = 4.75IGISDD65 pKa = 3.83AALTILSQTPQLVNEE80 pKa = 4.11AQAVGGLVNAAAGLAGGIAAFLEE103 pKa = 4.51PEE105 pKa = 4.11EE106 pKa = 4.74GNTGGVYY113 pKa = 10.3SQTSGSGWFGGFTWPPGAAAQPPLRR138 pKa = 11.84GPMDD142 pKa = 4.12AAVVPYY148 pKa = 10.82NEE150 pKa = 4.89LEE152 pKa = 4.04VGIPGIPDD160 pKa = 3.05WVLNMLPEE168 pKa = 4.7LPSLSDD174 pKa = 2.81IFSRR178 pKa = 11.84IAHH181 pKa = 6.85GIWTSYY187 pKa = 9.95YY188 pKa = 7.83NAGRR192 pKa = 11.84IVVQRR197 pKa = 11.84ALSSEE202 pKa = 4.11MQQLLRR208 pKa = 11.84DD209 pKa = 3.79LRR211 pKa = 11.84NGFSSTLQGIEE222 pKa = 4.1FSDD225 pKa = 4.01PVQAIANQIEE235 pKa = 4.62RR236 pKa = 11.84IQAQRR241 pKa = 11.84RR242 pKa = 11.84IDD244 pKa = 3.58NARR247 pKa = 11.84LLSYY251 pKa = 11.07AEE253 pKa = 4.21RR254 pKa = 11.84NQIDD258 pKa = 3.52VSNIVTALQEE268 pKa = 4.46GAQTAAGALRR278 pKa = 11.84DD279 pKa = 3.59AGGAVVGQVGNLAMDD294 pKa = 4.37IANLPIDD301 pKa = 4.94GYY303 pKa = 11.23NALSDD308 pKa = 3.69GVHH311 pKa = 7.05RR312 pKa = 11.84FGQWISISGPTGGTEE327 pKa = 4.16HH328 pKa = 6.99YY329 pKa = 10.86SFPDD333 pKa = 2.71WMLYY337 pKa = 10.43LLEE340 pKa = 4.3EE341 pKa = 4.55VEE343 pKa = 4.27TDD345 pKa = 3.93FTPRR349 pKa = 11.84VPLKK353 pKa = 10.77RR354 pKa = 11.84KK355 pKa = 10.12LEE357 pKa = 4.19EE358 pKa = 3.96NNGNQRR364 pKa = 11.84KK365 pKa = 9.15KK366 pKa = 10.5RR367 pKa = 11.84STGAKK372 pKa = 8.77TSSKK376 pKa = 9.46TSKK379 pKa = 8.86TSKK382 pKa = 9.94KK383 pKa = 9.83RR384 pKa = 11.84RR385 pKa = 11.84SS386 pKa = 3.28
MM1 pKa = 7.41GALLAIPEE9 pKa = 4.53IIAAFSIAGAEE20 pKa = 4.02ALEE23 pKa = 4.17ITGGSLAAIAAGEE36 pKa = 4.06GLAAAEE42 pKa = 4.51ALTEE46 pKa = 4.13SAALIGSEE54 pKa = 4.21ALTMSEE60 pKa = 4.75IGISDD65 pKa = 3.83AALTILSQTPQLVNEE80 pKa = 4.11AQAVGGLVNAAAGLAGGIAAFLEE103 pKa = 4.51PEE105 pKa = 4.11EE106 pKa = 4.74GNTGGVYY113 pKa = 10.3SQTSGSGWFGGFTWPPGAAAQPPLRR138 pKa = 11.84GPMDD142 pKa = 4.12AAVVPYY148 pKa = 10.82NEE150 pKa = 4.89LEE152 pKa = 4.04VGIPGIPDD160 pKa = 3.05WVLNMLPEE168 pKa = 4.7LPSLSDD174 pKa = 2.81IFSRR178 pKa = 11.84IAHH181 pKa = 6.85GIWTSYY187 pKa = 9.95YY188 pKa = 7.83NAGRR192 pKa = 11.84IVVQRR197 pKa = 11.84ALSSEE202 pKa = 4.11MQQLLRR208 pKa = 11.84DD209 pKa = 3.79LRR211 pKa = 11.84NGFSSTLQGIEE222 pKa = 4.1FSDD225 pKa = 4.01PVQAIANQIEE235 pKa = 4.62RR236 pKa = 11.84IQAQRR241 pKa = 11.84RR242 pKa = 11.84IDD244 pKa = 3.58NARR247 pKa = 11.84LLSYY251 pKa = 11.07AEE253 pKa = 4.21RR254 pKa = 11.84NQIDD258 pKa = 3.52VSNIVTALQEE268 pKa = 4.46GAQTAAGALRR278 pKa = 11.84DD279 pKa = 3.59AGGAVVGQVGNLAMDD294 pKa = 4.37IANLPIDD301 pKa = 4.94GYY303 pKa = 11.23NALSDD308 pKa = 3.69GVHH311 pKa = 7.05RR312 pKa = 11.84FGQWISISGPTGGTEE327 pKa = 4.16HH328 pKa = 6.99YY329 pKa = 10.86SFPDD333 pKa = 2.71WMLYY337 pKa = 10.43LLEE340 pKa = 4.3EE341 pKa = 4.55VEE343 pKa = 4.27TDD345 pKa = 3.93FTPRR349 pKa = 11.84VPLKK353 pKa = 10.77RR354 pKa = 11.84KK355 pKa = 10.12LEE357 pKa = 4.19EE358 pKa = 3.96NNGNQRR364 pKa = 11.84KK365 pKa = 9.15KK366 pKa = 10.5RR367 pKa = 11.84STGAKK372 pKa = 8.77TSSKK376 pKa = 9.46TSKK379 pKa = 8.86TSKK382 pKa = 9.94KK383 pKa = 9.83RR384 pKa = 11.84RR385 pKa = 11.84SS386 pKa = 3.28
Molecular weight: 40.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V0CLV8|A0A1V0CLV8_9POLY Major capsid protein VP1 OS=Vicugna pacos polyomavirus 1 OX=2169757 GN=VP1 PE=3 SV=1
MM1 pKa = 7.95DD2 pKa = 3.65RR3 pKa = 11.84TLDD6 pKa = 3.6RR7 pKa = 11.84EE8 pKa = 4.11EE9 pKa = 4.61SKK11 pKa = 11.33EE12 pKa = 3.91LMALLGLSMSCWGNIPMMRR31 pKa = 11.84RR32 pKa = 11.84AYY34 pKa = 9.44KK35 pKa = 8.07VQCRR39 pKa = 11.84NLHH42 pKa = 6.06PDD44 pKa = 2.89KK45 pKa = 11.51GGDD48 pKa = 3.52EE49 pKa = 4.33EE50 pKa = 4.56KK51 pKa = 10.29MKK53 pKa = 10.78RR54 pKa = 11.84LTEE57 pKa = 4.23LYY59 pKa = 10.15KK60 pKa = 10.81KK61 pKa = 10.77LEE63 pKa = 3.99EE64 pKa = 4.16TLGVIHH70 pKa = 6.28SQNEE74 pKa = 4.23SEE76 pKa = 4.14EE77 pKa = 4.55GSWNASEE84 pKa = 5.44VVLSDD89 pKa = 4.37PNCPCCQNLAGLNIGDD105 pKa = 3.67IYY107 pKa = 11.4GDD109 pKa = 3.39IFSEE113 pKa = 5.26LIVKK117 pKa = 9.99DD118 pKa = 3.46WLSCKK123 pKa = 10.43LGWNTKK129 pKa = 7.4CTCFMCKK136 pKa = 9.16LRR138 pKa = 11.84RR139 pKa = 11.84RR140 pKa = 11.84HH141 pKa = 5.53RR142 pKa = 11.84LRR144 pKa = 11.84VKK146 pKa = 10.13AYY148 pKa = 9.12KK149 pKa = 10.28RR150 pKa = 11.84PLKK153 pKa = 9.94WVEE156 pKa = 4.4CYY158 pKa = 10.62CFEE161 pKa = 5.83CYY163 pKa = 10.55VEE165 pKa = 4.11WFGFCRR171 pKa = 11.84TFEE174 pKa = 4.84ANFWWQKK181 pKa = 10.77IIFFTPFLEE190 pKa = 4.68IKK192 pKa = 10.39LL193 pKa = 3.95
MM1 pKa = 7.95DD2 pKa = 3.65RR3 pKa = 11.84TLDD6 pKa = 3.6RR7 pKa = 11.84EE8 pKa = 4.11EE9 pKa = 4.61SKK11 pKa = 11.33EE12 pKa = 3.91LMALLGLSMSCWGNIPMMRR31 pKa = 11.84RR32 pKa = 11.84AYY34 pKa = 9.44KK35 pKa = 8.07VQCRR39 pKa = 11.84NLHH42 pKa = 6.06PDD44 pKa = 2.89KK45 pKa = 11.51GGDD48 pKa = 3.52EE49 pKa = 4.33EE50 pKa = 4.56KK51 pKa = 10.29MKK53 pKa = 10.78RR54 pKa = 11.84LTEE57 pKa = 4.23LYY59 pKa = 10.15KK60 pKa = 10.81KK61 pKa = 10.77LEE63 pKa = 3.99EE64 pKa = 4.16TLGVIHH70 pKa = 6.28SQNEE74 pKa = 4.23SEE76 pKa = 4.14EE77 pKa = 4.55GSWNASEE84 pKa = 5.44VVLSDD89 pKa = 4.37PNCPCCQNLAGLNIGDD105 pKa = 3.67IYY107 pKa = 11.4GDD109 pKa = 3.39IFSEE113 pKa = 5.26LIVKK117 pKa = 9.99DD118 pKa = 3.46WLSCKK123 pKa = 10.43LGWNTKK129 pKa = 7.4CTCFMCKK136 pKa = 9.16LRR138 pKa = 11.84RR139 pKa = 11.84RR140 pKa = 11.84HH141 pKa = 5.53RR142 pKa = 11.84LRR144 pKa = 11.84VKK146 pKa = 10.13AYY148 pKa = 9.12KK149 pKa = 10.28RR150 pKa = 11.84PLKK153 pKa = 9.94WVEE156 pKa = 4.4CYY158 pKa = 10.62CFEE161 pKa = 5.83CYY163 pKa = 10.55VEE165 pKa = 4.11WFGFCRR171 pKa = 11.84TFEE174 pKa = 4.84ANFWWQKK181 pKa = 10.77IIFFTPFLEE190 pKa = 4.68IKK192 pKa = 10.39LL193 pKa = 3.95
Molecular weight: 22.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1850 |
193 |
657 |
370.0 |
41.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.432 ± 1.768 | 1.838 ± 0.828 |
4.649 ± 0.624 | 7.081 ± 0.592 |
3.73 ± 0.393 | 7.838 ± 1.26 |
1.459 ± 0.217 | 5.459 ± 0.586 |
5.946 ± 1.271 | 9.297 ± 0.399 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.757 ± 0.437 | 5.892 ± 0.506 |
4.973 ± 0.723 | 4.649 ± 0.491 |
4.919 ± 0.601 | 5.892 ± 0.782 |
5.514 ± 0.85 | 6.541 ± 1.045 |
1.784 ± 0.37 | 3.351 ± 0.408 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
