
Toxocara canis (Canine roundworm)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Spirurina; Ascaridomorpha; Ascaridoidea; Toxocaridae; Toxocara
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
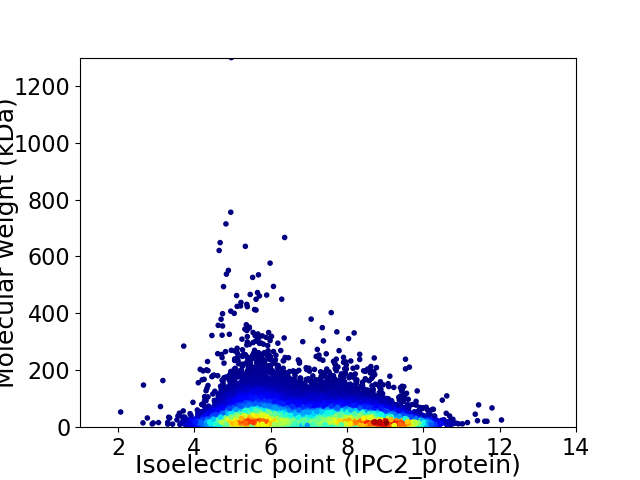
Virtual 2D-PAGE plot for 18499 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B2UV47|A0A0B2UV47_TOXCA UPF0046 protein C25E10.12 OS=Toxocara canis OX=6265 GN=C25E10.12 PE=4 SV=1
MM1 pKa = 7.16CTDD4 pKa = 2.86VDD6 pKa = 4.41MSLEE10 pKa = 4.11MCTDD14 pKa = 2.63VHH16 pKa = 6.43MSLEE20 pKa = 4.51TYY22 pKa = 10.33TDD24 pKa = 2.98VDD26 pKa = 3.64VSLEE30 pKa = 4.03VCTDD34 pKa = 2.77VDD36 pKa = 3.89MSLAMCTDD44 pKa = 3.04VDD46 pKa = 4.6VSLEE50 pKa = 4.07TCTDD54 pKa = 3.53VVVSLDD60 pKa = 3.17MCTDD64 pKa = 3.18VDD66 pKa = 4.7VSPDD70 pKa = 2.99MCTDD74 pKa = 2.88VDD76 pKa = 4.58VSLKK80 pKa = 9.75MCTDD84 pKa = 3.23VDD86 pKa = 4.26VSPDD90 pKa = 2.99MCTDD94 pKa = 2.88VDD96 pKa = 4.58VSLKK100 pKa = 9.75MCTDD104 pKa = 3.23VDD106 pKa = 4.26VSPDD110 pKa = 2.99MCTDD114 pKa = 2.88VDD116 pKa = 4.58VSLKK120 pKa = 9.75MCTDD124 pKa = 3.23VDD126 pKa = 4.26VSPDD130 pKa = 2.99MCTDD134 pKa = 2.88VDD136 pKa = 4.58VSLKK140 pKa = 9.75MCTDD144 pKa = 3.23VDD146 pKa = 4.26VSPDD150 pKa = 2.99MCTDD154 pKa = 2.88VDD156 pKa = 4.58VSLKK160 pKa = 9.75MCTDD164 pKa = 2.97VDD166 pKa = 4.19VSLKK170 pKa = 9.77MCTDD174 pKa = 2.8MDD176 pKa = 3.48VRR178 pKa = 11.84RR179 pKa = 11.84IQFSCDD185 pKa = 2.65VFLANDD191 pKa = 4.5FSNKK195 pKa = 6.85TLKK198 pKa = 10.69DD199 pKa = 3.58SLLMIVRR206 pKa = 11.84QRR208 pKa = 11.84INVEE212 pKa = 4.14FYY214 pKa = 11.04DD215 pKa = 3.98PFLLRR220 pKa = 11.84ILAEE224 pKa = 3.98NSLKK228 pKa = 10.79KK229 pKa = 10.55FF230 pKa = 3.55
MM1 pKa = 7.16CTDD4 pKa = 2.86VDD6 pKa = 4.41MSLEE10 pKa = 4.11MCTDD14 pKa = 2.63VHH16 pKa = 6.43MSLEE20 pKa = 4.51TYY22 pKa = 10.33TDD24 pKa = 2.98VDD26 pKa = 3.64VSLEE30 pKa = 4.03VCTDD34 pKa = 2.77VDD36 pKa = 3.89MSLAMCTDD44 pKa = 3.04VDD46 pKa = 4.6VSLEE50 pKa = 4.07TCTDD54 pKa = 3.53VVVSLDD60 pKa = 3.17MCTDD64 pKa = 3.18VDD66 pKa = 4.7VSPDD70 pKa = 2.99MCTDD74 pKa = 2.88VDD76 pKa = 4.58VSLKK80 pKa = 9.75MCTDD84 pKa = 3.23VDD86 pKa = 4.26VSPDD90 pKa = 2.99MCTDD94 pKa = 2.88VDD96 pKa = 4.58VSLKK100 pKa = 9.75MCTDD104 pKa = 3.23VDD106 pKa = 4.26VSPDD110 pKa = 2.99MCTDD114 pKa = 2.88VDD116 pKa = 4.58VSLKK120 pKa = 9.75MCTDD124 pKa = 3.23VDD126 pKa = 4.26VSPDD130 pKa = 2.99MCTDD134 pKa = 2.88VDD136 pKa = 4.58VSLKK140 pKa = 9.75MCTDD144 pKa = 3.23VDD146 pKa = 4.26VSPDD150 pKa = 2.99MCTDD154 pKa = 2.88VDD156 pKa = 4.58VSLKK160 pKa = 9.75MCTDD164 pKa = 2.97VDD166 pKa = 4.19VSLKK170 pKa = 9.77MCTDD174 pKa = 2.8MDD176 pKa = 3.48VRR178 pKa = 11.84RR179 pKa = 11.84IQFSCDD185 pKa = 2.65VFLANDD191 pKa = 4.5FSNKK195 pKa = 6.85TLKK198 pKa = 10.69DD199 pKa = 3.58SLLMIVRR206 pKa = 11.84QRR208 pKa = 11.84INVEE212 pKa = 4.14FYY214 pKa = 11.04DD215 pKa = 3.98PFLLRR220 pKa = 11.84ILAEE224 pKa = 3.98NSLKK228 pKa = 10.79KK229 pKa = 10.55FF230 pKa = 3.55
Molecular weight: 25.59 kDa
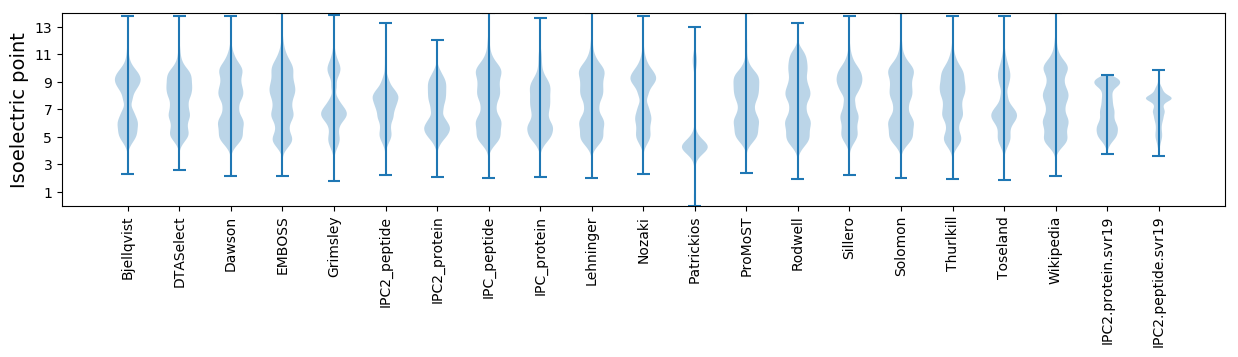
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B2W3E4|A0A0B2W3E4_TOXCA Uncharacterized protein OS=Toxocara canis OX=6265 GN=Tcan_12348 PE=4 SV=1
MM1 pKa = 7.3TGQRR5 pKa = 11.84SMTGQRR11 pKa = 11.84SMTGQRR17 pKa = 11.84SMTGQRR23 pKa = 11.84SMTGQRR29 pKa = 11.84SMTGQRR35 pKa = 11.84SMTGQRR41 pKa = 11.84SMTGQRR47 pKa = 11.84SMTGQRR53 pKa = 11.84SMTGQRR59 pKa = 11.84SMTGQRR65 pKa = 11.84SMTGQRR71 pKa = 11.84SMTGQRR77 pKa = 11.84SMTGQRR83 pKa = 11.84SMTGQRR89 pKa = 11.84SMTGQRR95 pKa = 11.84SMTGQRR101 pKa = 11.84SMTGQRR107 pKa = 11.84SMTGQRR113 pKa = 11.84SMTGQRR119 pKa = 11.84SMTGQRR125 pKa = 11.84SMTGQRR131 pKa = 11.84SMTGQRR137 pKa = 11.84SMTGQRR143 pKa = 11.84SMTGQRR149 pKa = 11.84SMTGQRR155 pKa = 11.84SMTGQRR161 pKa = 11.84SMTGQRR167 pKa = 11.84SMTGQRR173 pKa = 11.84SMTGQRR179 pKa = 11.84SMTGQRR185 pKa = 11.84SMTGQRR191 pKa = 11.84SMTGQRR197 pKa = 11.84SILLVIEE204 pKa = 4.49PSFF207 pKa = 3.69
MM1 pKa = 7.3TGQRR5 pKa = 11.84SMTGQRR11 pKa = 11.84SMTGQRR17 pKa = 11.84SMTGQRR23 pKa = 11.84SMTGQRR29 pKa = 11.84SMTGQRR35 pKa = 11.84SMTGQRR41 pKa = 11.84SMTGQRR47 pKa = 11.84SMTGQRR53 pKa = 11.84SMTGQRR59 pKa = 11.84SMTGQRR65 pKa = 11.84SMTGQRR71 pKa = 11.84SMTGQRR77 pKa = 11.84SMTGQRR83 pKa = 11.84SMTGQRR89 pKa = 11.84SMTGQRR95 pKa = 11.84SMTGQRR101 pKa = 11.84SMTGQRR107 pKa = 11.84SMTGQRR113 pKa = 11.84SMTGQRR119 pKa = 11.84SMTGQRR125 pKa = 11.84SMTGQRR131 pKa = 11.84SMTGQRR137 pKa = 11.84SMTGQRR143 pKa = 11.84SMTGQRR149 pKa = 11.84SMTGQRR155 pKa = 11.84SMTGQRR161 pKa = 11.84SMTGQRR167 pKa = 11.84SMTGQRR173 pKa = 11.84SMTGQRR179 pKa = 11.84SMTGQRR185 pKa = 11.84SMTGQRR191 pKa = 11.84SMTGQRR197 pKa = 11.84SILLVIEE204 pKa = 4.49PSFF207 pKa = 3.69
Molecular weight: 22.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7137888 |
24 |
11591 |
385.9 |
43.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.537 ± 0.019 | 2.283 ± 0.018 |
5.221 ± 0.015 | 6.599 ± 0.027 |
4.056 ± 0.016 | 5.661 ± 0.028 |
2.413 ± 0.01 | 5.382 ± 0.016 |
5.361 ± 0.019 | 8.932 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.532 ± 0.01 | 4.194 ± 0.014 |
4.854 ± 0.026 | 3.963 ± 0.016 |
6.435 ± 0.019 | 8.368 ± 0.026 |
5.583 ± 0.021 | 6.57 ± 0.024 |
1.14 ± 0.009 | 2.882 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
