
Luteimonas sp. JM171
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Luteimonas; unclassified Luteimonas
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
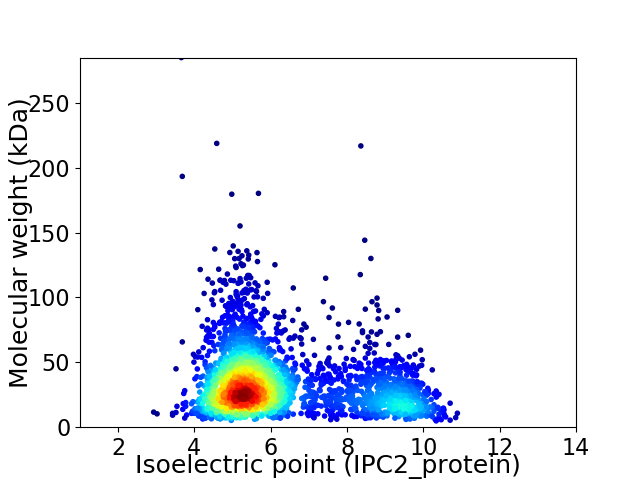
Virtual 2D-PAGE plot for 2668 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B3W5G1|A0A1B3W5G1_9GAMM Uncharacterized protein OS=Luteimonas sp. JM171 OX=1896164 GN=BGP89_02155 PE=4 SV=1
AA1 pKa = 7.7EE2 pKa = 5.0GVDD5 pKa = 3.49STAIGFFSHH14 pKa = 6.92AEE16 pKa = 3.86AAGSVALGVDD26 pKa = 3.77SVADD30 pKa = 3.98RR31 pKa = 11.84EE32 pKa = 4.48NTVSVGSAGNEE43 pKa = 3.49RR44 pKa = 11.84QIANVADD51 pKa = 3.79GTEE54 pKa = 3.97ATDD57 pKa = 4.38AVNLGQLEE65 pKa = 4.34AVTSDD70 pKa = 2.84THH72 pKa = 6.84YY73 pKa = 10.66FAATGDD79 pKa = 3.88GEE81 pKa = 4.66AFATGLDD88 pKa = 3.68ATASGSDD95 pKa = 3.18AFAEE99 pKa = 4.48GDD101 pKa = 3.66YY102 pKa = 10.51STATGSASWALGEE115 pKa = 4.21GASAFGSGAQAWGQYY130 pKa = 7.99ATASGFNAYY139 pKa = 10.26AEE141 pKa = 4.78GDD143 pKa = 3.65STTAVGGWLYY153 pKa = 11.35YY154 pKa = 10.23EE155 pKa = 5.26DD156 pKa = 5.04PATGDD161 pKa = 3.57VLLDD165 pKa = 3.34ATTTAFEE172 pKa = 4.35VGASAFGAGAQALGAFSTASGAAAEE197 pKa = 4.43AGGLQATATGYY208 pKa = 10.69SSSASGNYY216 pKa = 7.86STASGGFSSAGDD228 pKa = 3.53VGATALGYY236 pKa = 10.56GADD239 pKa = 3.7ASAGYY244 pKa = 8.37ATAVGTAAWAQGVSSVAVGEE264 pKa = 4.13YY265 pKa = 10.6SEE267 pKa = 4.34ATGAEE272 pKa = 4.32SVAVGGTVFGFIPSRR287 pKa = 11.84ASGAGAAAFGAGAWATGDD305 pKa = 3.39EE306 pKa = 4.5STALGWISSADD317 pKa = 3.41GDD319 pKa = 3.97YY320 pKa = 10.37STAVGAASWAEE331 pKa = 3.95AADD334 pKa = 3.93STVVGYY340 pKa = 10.5GGYY343 pKa = 10.77ASAEE347 pKa = 3.91NSVALGAGSVADD359 pKa = 4.16RR360 pKa = 11.84ADD362 pKa = 3.72SVSVGSAGGEE372 pKa = 3.67RR373 pKa = 11.84QITNVAAGTEE383 pKa = 4.26GTDD386 pKa = 3.31AVNVEE391 pKa = 4.17QLTDD395 pKa = 4.71AIAAVDD401 pKa = 3.68TEE403 pKa = 4.35GNRR406 pKa = 11.84FFDD409 pKa = 3.5ASAGVDD415 pKa = 3.54EE416 pKa = 5.5AGVGASADD424 pKa = 3.68GEE426 pKa = 4.48YY427 pKa = 9.98ATAGGEE433 pKa = 4.03LASASGDD440 pKa = 3.32LSVAMGALSDD450 pKa = 3.69ASGWGATAVGVEE462 pKa = 4.61SVASGEE468 pKa = 3.83LAAAFGGAAEE478 pKa = 4.29AFGDD482 pKa = 4.44FSTALGGFSTAAGYY496 pKa = 11.09NSTALGNFATATADD510 pKa = 3.3NSVALGADD518 pKa = 3.38SVADD522 pKa = 3.92RR523 pKa = 11.84DD524 pKa = 3.91NTVSVGSEE532 pKa = 3.65GSEE535 pKa = 3.59RR536 pKa = 11.84QITNVAAGTEE546 pKa = 4.23GTDD549 pKa = 4.02AVNLGQLEE557 pKa = 4.09EE558 pKa = 4.05ATAYY562 pKa = 9.64NQYY565 pKa = 10.09FRR567 pKa = 11.84ATGEE571 pKa = 4.13GEE573 pKa = 3.64AWAVGEE579 pKa = 4.55DD580 pKa = 3.34ATAAGSNAWAGGDD593 pKa = 3.47YY594 pKa = 9.68STAVGSTAEE603 pKa = 3.82AWMEE607 pKa = 3.89GASAFGSGAFALEE620 pKa = 4.18EE621 pKa = 3.95NATALGFNTVAGGIDD636 pKa = 3.6AVALGSYY643 pKa = 10.43AEE645 pKa = 4.6AYY647 pKa = 9.98GEE649 pKa = 3.84QSIALGSQAYY659 pKa = 9.45AAEE662 pKa = 4.46TGASAIGAGAVAEE675 pKa = 4.55GAYY678 pKa = 8.92STATGSEE685 pKa = 3.84ASASGLQGTATGFRR699 pKa = 11.84SEE701 pKa = 3.97ASGDD705 pKa = 3.14GSAAYY710 pKa = 9.75GGYY713 pKa = 9.98SVAQGFGASALGYY726 pKa = 9.49GANALADD733 pKa = 3.78GATAVGFAADD743 pKa = 3.56ASGEE747 pKa = 4.16SSVAVGEE754 pKa = 4.05AAIASGFNSVAVGGASFFGLLPTEE778 pKa = 4.13ATGDD782 pKa = 3.62YY783 pKa = 8.57ATAVGAGSWAPGANGTALGSLSMATGVDD811 pKa = 3.74SLAVGADD818 pKa = 3.2AFASAEE824 pKa = 4.13GAVALGVGSEE834 pKa = 3.91ADD836 pKa = 3.3RR837 pKa = 11.84DD838 pKa = 3.94YY839 pKa = 11.58SVSVGSAGNEE849 pKa = 3.59RR850 pKa = 11.84QITNVAAGTEE860 pKa = 4.1ATDD863 pKa = 4.26AVNLGQLEE871 pKa = 4.38AVTGDD876 pKa = 3.34THH878 pKa = 6.85YY879 pKa = 10.57FAASGEE885 pKa = 4.22GQAWATGDD893 pKa = 3.75DD894 pKa = 3.84ATASGSDD901 pKa = 3.29AFAEE905 pKa = 4.72GDD907 pKa = 3.43FSTATGATSFAVGQGASAFGSGAQAWGQYY936 pKa = 7.97ATASGYY942 pKa = 10.39NAVAEE947 pKa = 4.79GDD949 pKa = 3.51SSTAVGGWLYY959 pKa = 11.35YY960 pKa = 10.19EE961 pKa = 5.18DD962 pKa = 4.72PATGEE967 pKa = 4.15VLLDD971 pKa = 3.41QTTTALEE978 pKa = 4.32AGASAFGAGAQALGAFSTATGAASTAEE1005 pKa = 4.17GLQATATGYY1014 pKa = 11.04SSTASGLASTANGGFSEE1031 pKa = 4.32ATGEE1035 pKa = 3.95FSTALGYY1042 pKa = 10.7GAVASNTRR1050 pKa = 11.84TTAVGISAEE1059 pKa = 3.98ASGLNATAVGNTATASGSAGVAVGSSSSASGVNGTAVGGTAWATGEE1105 pKa = 4.2DD1106 pKa = 3.93ATALGQFAWATRR1118 pKa = 11.84AGATALGQDD1127 pKa = 3.44TFAYY1131 pKa = 10.36ADD1133 pKa = 3.55GATAVGASAWSNGVNSVALGSGSRR1157 pKa = 11.84ASADD1161 pKa = 3.07NVVSVGNGTGNSGFPATRR1179 pKa = 11.84RR1180 pKa = 11.84IVNVSDD1186 pKa = 4.34GVDD1189 pKa = 3.64DD1190 pKa = 4.17NDD1192 pKa = 3.89AVNVGQMGEE1201 pKa = 4.38FVAEE1205 pKa = 4.38ANRR1208 pKa = 11.84HH1209 pKa = 4.12FQATGEE1215 pKa = 3.99ADD1217 pKa = 3.32AYY1219 pKa = 10.92ALGEE1223 pKa = 4.09EE1224 pKa = 4.56STAAGANAYY1233 pKa = 10.21AEE1235 pKa = 4.65ADD1237 pKa = 3.57YY1238 pKa = 9.9STAVGSASSAFEE1250 pKa = 4.27LGASAFGSGALAGQQQATAVGFNASAAGSNSSAFGAGADD1289 pKa = 3.74AYY1291 pKa = 10.31GDD1293 pKa = 3.41ASTAVGGQGEE1303 pKa = 4.37AFDD1306 pKa = 3.92EE1307 pKa = 4.79FGWPIEE1313 pKa = 4.21DD1314 pKa = 4.14SEE1316 pKa = 4.66GNPVLVAATAADD1328 pKa = 3.74RR1329 pKa = 11.84GTAVGAGAQSTGTGSASLGYY1349 pKa = 9.45GARR1352 pKa = 11.84AEE1354 pKa = 4.7GEE1356 pKa = 3.81FSLAGGHH1363 pKa = 6.53KK1364 pKa = 9.93AHH1366 pKa = 6.73AQGGYY1371 pKa = 7.59TVSLGDD1377 pKa = 3.44NSRR1380 pKa = 11.84ANGDD1384 pKa = 3.12QSTATGSYY1392 pKa = 10.02AWATEE1397 pKa = 4.14SGSTALGSFAWATGEE1412 pKa = 3.8EE1413 pKa = 4.19ATAVGVDD1420 pKa = 3.65SYY1422 pKa = 11.44ATGRR1426 pKa = 11.84RR1427 pKa = 11.84SVSIGSGSWSPGVSSVAVGRR1447 pKa = 11.84GAFGYY1452 pKa = 10.59DD1453 pKa = 3.83DD1454 pKa = 3.58YY1455 pKa = 11.44TVAVGHH1461 pKa = 5.8QALGNDD1467 pKa = 2.75IHH1469 pKa = 7.94AIAVGTNSLANAEE1482 pKa = 4.15SAIAIGAGATVAATAPNSVALGSGSVAEE1510 pKa = 4.39RR1511 pKa = 11.84EE1512 pKa = 4.19RR1513 pKa = 11.84TLSVGSVGNEE1523 pKa = 3.43RR1524 pKa = 11.84QITNVAAGTQDD1535 pKa = 3.01TDD1537 pKa = 3.82AVNLGQLNAVADD1549 pKa = 4.51GVTALADD1556 pKa = 3.6SAVVYY1561 pKa = 10.87DD1562 pKa = 4.42DD1563 pKa = 4.75DD1564 pKa = 4.46SRR1566 pKa = 11.84GAITLAGEE1574 pKa = 4.2EE1575 pKa = 4.52GTLVTNLADD1584 pKa = 3.83GEE1586 pKa = 4.51VSAEE1590 pKa = 4.06STDD1593 pKa = 3.51AVTGSQLFEE1602 pKa = 4.01TNEE1605 pKa = 3.87RR1606 pKa = 11.84VGVVEE1611 pKa = 5.01GRR1613 pKa = 11.84VDD1615 pKa = 3.76DD1616 pKa = 4.35LDD1618 pKa = 3.81VRR1620 pKa = 11.84IGDD1623 pKa = 3.91VEE1625 pKa = 4.5GMASNAISYY1634 pKa = 10.74DD1635 pKa = 3.5DD1636 pKa = 4.65DD1637 pKa = 3.83SRR1639 pKa = 11.84AVATLDD1645 pKa = 3.64GEE1647 pKa = 4.65GGTRR1651 pKa = 11.84LTNLAPGLIRR1661 pKa = 11.84NGSSDD1666 pKa = 3.78AVTGGQVFASMGTIATALGGGTVVDD1691 pKa = 3.97ANGRR1695 pKa = 11.84LIGTVYY1701 pKa = 10.46SVQGNYY1707 pKa = 9.66YY1708 pKa = 9.87GNVGEE1713 pKa = 5.47ALAALDD1719 pKa = 3.44THH1721 pKa = 7.72LEE1723 pKa = 4.21GFGNRR1728 pKa = 11.84LSGVEE1733 pKa = 4.13AIVRR1737 pKa = 11.84SAQGPGNGRR1746 pKa = 11.84VAVGGAGRR1754 pKa = 11.84AQLGAEE1760 pKa = 5.09DD1761 pKa = 4.2INAIAAGDD1769 pKa = 3.65DD1770 pKa = 4.09AYY1772 pKa = 11.05AHH1774 pKa = 6.67GPNDD1778 pKa = 3.21IAIGGNAKK1786 pKa = 10.03VNADD1790 pKa = 2.9GSTAVGSNATITADD1804 pKa = 3.1ATNAVAIGEE1813 pKa = 4.43GTSVSAASGTAVGQGASVTAEE1834 pKa = 3.88GAVALGQGSVADD1846 pKa = 4.17RR1847 pKa = 11.84ANTVSVGAEE1856 pKa = 3.68GSEE1859 pKa = 3.79RR1860 pKa = 11.84QIVNVAAGTADD1871 pKa = 3.25TDD1873 pKa = 3.81AANVGQVNAAAAQSRR1888 pKa = 11.84SYY1890 pKa = 10.24TDD1892 pKa = 3.31TVATQTLSSANAYY1905 pKa = 8.31TDD1907 pKa = 3.12QQFAVWNDD1915 pKa = 2.76NFEE1918 pKa = 4.27AFRR1921 pKa = 11.84GDD1923 pKa = 3.47VEE1925 pKa = 5.05RR1926 pKa = 11.84RR1927 pKa = 11.84FHH1929 pKa = 7.47DD1930 pKa = 3.02VDD1932 pKa = 4.0RR1933 pKa = 11.84RR1934 pKa = 11.84LDD1936 pKa = 3.47RR1937 pKa = 11.84QGAMSAAMLNMATSAAGIRR1956 pKa = 11.84TQNRR1960 pKa = 11.84IGVGVGFQSGEE1971 pKa = 4.01SALSVGYY1978 pKa = 9.93QRR1980 pKa = 11.84AISEE1984 pKa = 4.23RR1985 pKa = 11.84ATITFGGAFSGDD1997 pKa = 3.53EE1998 pKa = 4.05SSVGIGAGFGWW2009 pKa = 3.8
AA1 pKa = 7.7EE2 pKa = 5.0GVDD5 pKa = 3.49STAIGFFSHH14 pKa = 6.92AEE16 pKa = 3.86AAGSVALGVDD26 pKa = 3.77SVADD30 pKa = 3.98RR31 pKa = 11.84EE32 pKa = 4.48NTVSVGSAGNEE43 pKa = 3.49RR44 pKa = 11.84QIANVADD51 pKa = 3.79GTEE54 pKa = 3.97ATDD57 pKa = 4.38AVNLGQLEE65 pKa = 4.34AVTSDD70 pKa = 2.84THH72 pKa = 6.84YY73 pKa = 10.66FAATGDD79 pKa = 3.88GEE81 pKa = 4.66AFATGLDD88 pKa = 3.68ATASGSDD95 pKa = 3.18AFAEE99 pKa = 4.48GDD101 pKa = 3.66YY102 pKa = 10.51STATGSASWALGEE115 pKa = 4.21GASAFGSGAQAWGQYY130 pKa = 7.99ATASGFNAYY139 pKa = 10.26AEE141 pKa = 4.78GDD143 pKa = 3.65STTAVGGWLYY153 pKa = 11.35YY154 pKa = 10.23EE155 pKa = 5.26DD156 pKa = 5.04PATGDD161 pKa = 3.57VLLDD165 pKa = 3.34ATTTAFEE172 pKa = 4.35VGASAFGAGAQALGAFSTASGAAAEE197 pKa = 4.43AGGLQATATGYY208 pKa = 10.69SSSASGNYY216 pKa = 7.86STASGGFSSAGDD228 pKa = 3.53VGATALGYY236 pKa = 10.56GADD239 pKa = 3.7ASAGYY244 pKa = 8.37ATAVGTAAWAQGVSSVAVGEE264 pKa = 4.13YY265 pKa = 10.6SEE267 pKa = 4.34ATGAEE272 pKa = 4.32SVAVGGTVFGFIPSRR287 pKa = 11.84ASGAGAAAFGAGAWATGDD305 pKa = 3.39EE306 pKa = 4.5STALGWISSADD317 pKa = 3.41GDD319 pKa = 3.97YY320 pKa = 10.37STAVGAASWAEE331 pKa = 3.95AADD334 pKa = 3.93STVVGYY340 pKa = 10.5GGYY343 pKa = 10.77ASAEE347 pKa = 3.91NSVALGAGSVADD359 pKa = 4.16RR360 pKa = 11.84ADD362 pKa = 3.72SVSVGSAGGEE372 pKa = 3.67RR373 pKa = 11.84QITNVAAGTEE383 pKa = 4.26GTDD386 pKa = 3.31AVNVEE391 pKa = 4.17QLTDD395 pKa = 4.71AIAAVDD401 pKa = 3.68TEE403 pKa = 4.35GNRR406 pKa = 11.84FFDD409 pKa = 3.5ASAGVDD415 pKa = 3.54EE416 pKa = 5.5AGVGASADD424 pKa = 3.68GEE426 pKa = 4.48YY427 pKa = 9.98ATAGGEE433 pKa = 4.03LASASGDD440 pKa = 3.32LSVAMGALSDD450 pKa = 3.69ASGWGATAVGVEE462 pKa = 4.61SVASGEE468 pKa = 3.83LAAAFGGAAEE478 pKa = 4.29AFGDD482 pKa = 4.44FSTALGGFSTAAGYY496 pKa = 11.09NSTALGNFATATADD510 pKa = 3.3NSVALGADD518 pKa = 3.38SVADD522 pKa = 3.92RR523 pKa = 11.84DD524 pKa = 3.91NTVSVGSEE532 pKa = 3.65GSEE535 pKa = 3.59RR536 pKa = 11.84QITNVAAGTEE546 pKa = 4.23GTDD549 pKa = 4.02AVNLGQLEE557 pKa = 4.09EE558 pKa = 4.05ATAYY562 pKa = 9.64NQYY565 pKa = 10.09FRR567 pKa = 11.84ATGEE571 pKa = 4.13GEE573 pKa = 3.64AWAVGEE579 pKa = 4.55DD580 pKa = 3.34ATAAGSNAWAGGDD593 pKa = 3.47YY594 pKa = 9.68STAVGSTAEE603 pKa = 3.82AWMEE607 pKa = 3.89GASAFGSGAFALEE620 pKa = 4.18EE621 pKa = 3.95NATALGFNTVAGGIDD636 pKa = 3.6AVALGSYY643 pKa = 10.43AEE645 pKa = 4.6AYY647 pKa = 9.98GEE649 pKa = 3.84QSIALGSQAYY659 pKa = 9.45AAEE662 pKa = 4.46TGASAIGAGAVAEE675 pKa = 4.55GAYY678 pKa = 8.92STATGSEE685 pKa = 3.84ASASGLQGTATGFRR699 pKa = 11.84SEE701 pKa = 3.97ASGDD705 pKa = 3.14GSAAYY710 pKa = 9.75GGYY713 pKa = 9.98SVAQGFGASALGYY726 pKa = 9.49GANALADD733 pKa = 3.78GATAVGFAADD743 pKa = 3.56ASGEE747 pKa = 4.16SSVAVGEE754 pKa = 4.05AAIASGFNSVAVGGASFFGLLPTEE778 pKa = 4.13ATGDD782 pKa = 3.62YY783 pKa = 8.57ATAVGAGSWAPGANGTALGSLSMATGVDD811 pKa = 3.74SLAVGADD818 pKa = 3.2AFASAEE824 pKa = 4.13GAVALGVGSEE834 pKa = 3.91ADD836 pKa = 3.3RR837 pKa = 11.84DD838 pKa = 3.94YY839 pKa = 11.58SVSVGSAGNEE849 pKa = 3.59RR850 pKa = 11.84QITNVAAGTEE860 pKa = 4.1ATDD863 pKa = 4.26AVNLGQLEE871 pKa = 4.38AVTGDD876 pKa = 3.34THH878 pKa = 6.85YY879 pKa = 10.57FAASGEE885 pKa = 4.22GQAWATGDD893 pKa = 3.75DD894 pKa = 3.84ATASGSDD901 pKa = 3.29AFAEE905 pKa = 4.72GDD907 pKa = 3.43FSTATGATSFAVGQGASAFGSGAQAWGQYY936 pKa = 7.97ATASGYY942 pKa = 10.39NAVAEE947 pKa = 4.79GDD949 pKa = 3.51SSTAVGGWLYY959 pKa = 11.35YY960 pKa = 10.19EE961 pKa = 5.18DD962 pKa = 4.72PATGEE967 pKa = 4.15VLLDD971 pKa = 3.41QTTTALEE978 pKa = 4.32AGASAFGAGAQALGAFSTATGAASTAEE1005 pKa = 4.17GLQATATGYY1014 pKa = 11.04SSTASGLASTANGGFSEE1031 pKa = 4.32ATGEE1035 pKa = 3.95FSTALGYY1042 pKa = 10.7GAVASNTRR1050 pKa = 11.84TTAVGISAEE1059 pKa = 3.98ASGLNATAVGNTATASGSAGVAVGSSSSASGVNGTAVGGTAWATGEE1105 pKa = 4.2DD1106 pKa = 3.93ATALGQFAWATRR1118 pKa = 11.84AGATALGQDD1127 pKa = 3.44TFAYY1131 pKa = 10.36ADD1133 pKa = 3.55GATAVGASAWSNGVNSVALGSGSRR1157 pKa = 11.84ASADD1161 pKa = 3.07NVVSVGNGTGNSGFPATRR1179 pKa = 11.84RR1180 pKa = 11.84IVNVSDD1186 pKa = 4.34GVDD1189 pKa = 3.64DD1190 pKa = 4.17NDD1192 pKa = 3.89AVNVGQMGEE1201 pKa = 4.38FVAEE1205 pKa = 4.38ANRR1208 pKa = 11.84HH1209 pKa = 4.12FQATGEE1215 pKa = 3.99ADD1217 pKa = 3.32AYY1219 pKa = 10.92ALGEE1223 pKa = 4.09EE1224 pKa = 4.56STAAGANAYY1233 pKa = 10.21AEE1235 pKa = 4.65ADD1237 pKa = 3.57YY1238 pKa = 9.9STAVGSASSAFEE1250 pKa = 4.27LGASAFGSGALAGQQQATAVGFNASAAGSNSSAFGAGADD1289 pKa = 3.74AYY1291 pKa = 10.31GDD1293 pKa = 3.41ASTAVGGQGEE1303 pKa = 4.37AFDD1306 pKa = 3.92EE1307 pKa = 4.79FGWPIEE1313 pKa = 4.21DD1314 pKa = 4.14SEE1316 pKa = 4.66GNPVLVAATAADD1328 pKa = 3.74RR1329 pKa = 11.84GTAVGAGAQSTGTGSASLGYY1349 pKa = 9.45GARR1352 pKa = 11.84AEE1354 pKa = 4.7GEE1356 pKa = 3.81FSLAGGHH1363 pKa = 6.53KK1364 pKa = 9.93AHH1366 pKa = 6.73AQGGYY1371 pKa = 7.59TVSLGDD1377 pKa = 3.44NSRR1380 pKa = 11.84ANGDD1384 pKa = 3.12QSTATGSYY1392 pKa = 10.02AWATEE1397 pKa = 4.14SGSTALGSFAWATGEE1412 pKa = 3.8EE1413 pKa = 4.19ATAVGVDD1420 pKa = 3.65SYY1422 pKa = 11.44ATGRR1426 pKa = 11.84RR1427 pKa = 11.84SVSIGSGSWSPGVSSVAVGRR1447 pKa = 11.84GAFGYY1452 pKa = 10.59DD1453 pKa = 3.83DD1454 pKa = 3.58YY1455 pKa = 11.44TVAVGHH1461 pKa = 5.8QALGNDD1467 pKa = 2.75IHH1469 pKa = 7.94AIAVGTNSLANAEE1482 pKa = 4.15SAIAIGAGATVAATAPNSVALGSGSVAEE1510 pKa = 4.39RR1511 pKa = 11.84EE1512 pKa = 4.19RR1513 pKa = 11.84TLSVGSVGNEE1523 pKa = 3.43RR1524 pKa = 11.84QITNVAAGTQDD1535 pKa = 3.01TDD1537 pKa = 3.82AVNLGQLNAVADD1549 pKa = 4.51GVTALADD1556 pKa = 3.6SAVVYY1561 pKa = 10.87DD1562 pKa = 4.42DD1563 pKa = 4.75DD1564 pKa = 4.46SRR1566 pKa = 11.84GAITLAGEE1574 pKa = 4.2EE1575 pKa = 4.52GTLVTNLADD1584 pKa = 3.83GEE1586 pKa = 4.51VSAEE1590 pKa = 4.06STDD1593 pKa = 3.51AVTGSQLFEE1602 pKa = 4.01TNEE1605 pKa = 3.87RR1606 pKa = 11.84VGVVEE1611 pKa = 5.01GRR1613 pKa = 11.84VDD1615 pKa = 3.76DD1616 pKa = 4.35LDD1618 pKa = 3.81VRR1620 pKa = 11.84IGDD1623 pKa = 3.91VEE1625 pKa = 4.5GMASNAISYY1634 pKa = 10.74DD1635 pKa = 3.5DD1636 pKa = 4.65DD1637 pKa = 3.83SRR1639 pKa = 11.84AVATLDD1645 pKa = 3.64GEE1647 pKa = 4.65GGTRR1651 pKa = 11.84LTNLAPGLIRR1661 pKa = 11.84NGSSDD1666 pKa = 3.78AVTGGQVFASMGTIATALGGGTVVDD1691 pKa = 3.97ANGRR1695 pKa = 11.84LIGTVYY1701 pKa = 10.46SVQGNYY1707 pKa = 9.66YY1708 pKa = 9.87GNVGEE1713 pKa = 5.47ALAALDD1719 pKa = 3.44THH1721 pKa = 7.72LEE1723 pKa = 4.21GFGNRR1728 pKa = 11.84LSGVEE1733 pKa = 4.13AIVRR1737 pKa = 11.84SAQGPGNGRR1746 pKa = 11.84VAVGGAGRR1754 pKa = 11.84AQLGAEE1760 pKa = 5.09DD1761 pKa = 4.2INAIAAGDD1769 pKa = 3.65DD1770 pKa = 4.09AYY1772 pKa = 11.05AHH1774 pKa = 6.67GPNDD1778 pKa = 3.21IAIGGNAKK1786 pKa = 10.03VNADD1790 pKa = 2.9GSTAVGSNATITADD1804 pKa = 3.1ATNAVAIGEE1813 pKa = 4.43GTSVSAASGTAVGQGASVTAEE1834 pKa = 3.88GAVALGQGSVADD1846 pKa = 4.17RR1847 pKa = 11.84ANTVSVGAEE1856 pKa = 3.68GSEE1859 pKa = 3.79RR1860 pKa = 11.84QIVNVAAGTADD1871 pKa = 3.25TDD1873 pKa = 3.81AANVGQVNAAAAQSRR1888 pKa = 11.84SYY1890 pKa = 10.24TDD1892 pKa = 3.31TVATQTLSSANAYY1905 pKa = 8.31TDD1907 pKa = 3.12QQFAVWNDD1915 pKa = 2.76NFEE1918 pKa = 4.27AFRR1921 pKa = 11.84GDD1923 pKa = 3.47VEE1925 pKa = 5.05RR1926 pKa = 11.84RR1927 pKa = 11.84FHH1929 pKa = 7.47DD1930 pKa = 3.02VDD1932 pKa = 4.0RR1933 pKa = 11.84RR1934 pKa = 11.84LDD1936 pKa = 3.47RR1937 pKa = 11.84QGAMSAAMLNMATSAAGIRR1956 pKa = 11.84TQNRR1960 pKa = 11.84IGVGVGFQSGEE1971 pKa = 4.01SALSVGYY1978 pKa = 9.93QRR1980 pKa = 11.84AISEE1984 pKa = 4.23RR1985 pKa = 11.84ATITFGGAFSGDD1997 pKa = 3.53EE1998 pKa = 4.05SSVGIGAGFGWW2009 pKa = 3.8
Molecular weight: 193.25 kDa
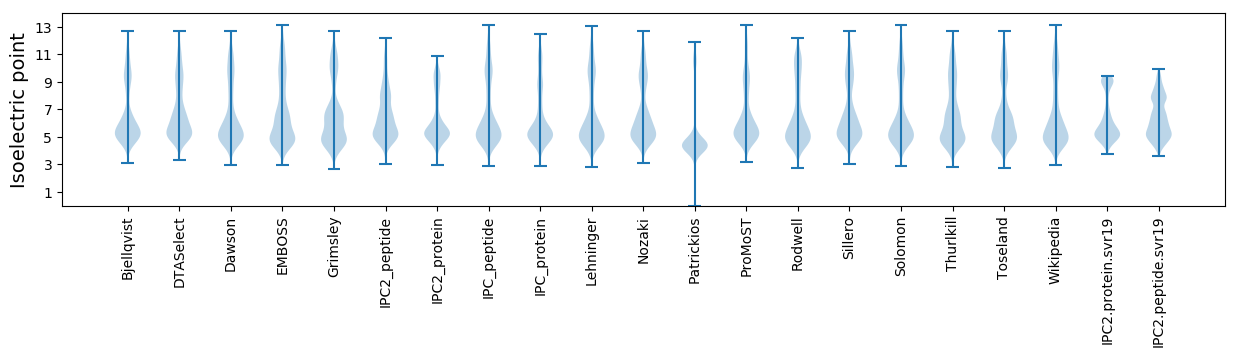
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B3W7M5|A0A1B3W7M5_9GAMM Uncharacterized protein OS=Luteimonas sp. JM171 OX=1896164 GN=BGP89_06615 PE=4 SV=1
MM1 pKa = 7.29HH2 pKa = 7.3HH3 pKa = 7.24PSASPEE9 pKa = 4.11SRR11 pKa = 11.84FLRR14 pKa = 11.84QALAGAAFLGLVAVSTLSAARR35 pKa = 11.84GGGPVGALALWLPGLPLLALAALALAGSRR64 pKa = 11.84GHH66 pKa = 7.07RR67 pKa = 11.84DD68 pKa = 3.27APASRR73 pKa = 11.84PAPASSRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84ARR84 pKa = 11.84PAQARR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84APARR95 pKa = 11.84PAHH98 pKa = 5.78PRR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84AAAA105 pKa = 3.68
MM1 pKa = 7.29HH2 pKa = 7.3HH3 pKa = 7.24PSASPEE9 pKa = 4.11SRR11 pKa = 11.84FLRR14 pKa = 11.84QALAGAAFLGLVAVSTLSAARR35 pKa = 11.84GGGPVGALALWLPGLPLLALAALALAGSRR64 pKa = 11.84GHH66 pKa = 7.07RR67 pKa = 11.84DD68 pKa = 3.27APASRR73 pKa = 11.84PAPASSRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84ARR84 pKa = 11.84PAQARR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84APARR95 pKa = 11.84PAHH98 pKa = 5.78PRR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84AAAA105 pKa = 3.68
Molecular weight: 10.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
882712 |
41 |
2925 |
330.9 |
35.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.072 ± 0.076 | 0.744 ± 0.013 |
6.011 ± 0.038 | 6.122 ± 0.049 |
3.301 ± 0.027 | 9.132 ± 0.05 |
2.192 ± 0.025 | 4.068 ± 0.039 |
2.129 ± 0.042 | 10.601 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.315 ± 0.023 | 2.282 ± 0.032 |
5.527 ± 0.037 | 3.544 ± 0.027 |
8.205 ± 0.055 | 4.931 ± 0.045 |
4.553 ± 0.036 | 7.595 ± 0.043 |
1.54 ± 0.021 | 2.138 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
