
Anaerosacchriphilus polymeriproducens
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Anaerosacchariphilus
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
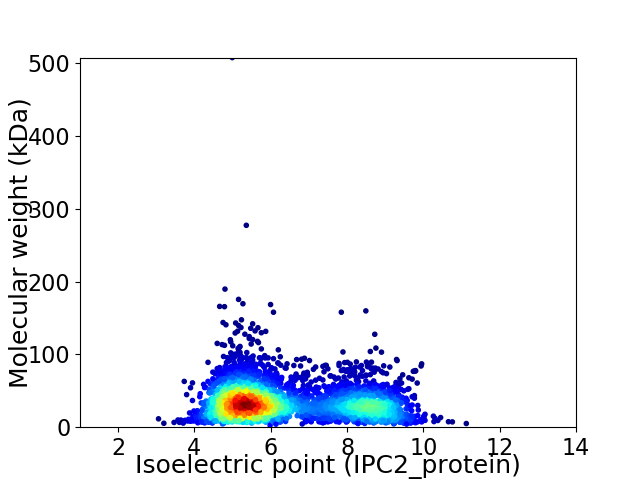
Virtual 2D-PAGE plot for 3531 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A371ASX5|A0A371ASX5_9FIRM Heme chaperone HemW OS=Anaerosacchriphilus polymeriproducens OX=1812858 GN=DWV06_14340 PE=3 SV=1
MM1 pKa = 7.61TNYY4 pKa = 10.26QSIFQTGTLEE14 pKa = 5.06SGFTYY19 pKa = 10.36NAPQVVDD26 pKa = 3.28NTSYY30 pKa = 11.53NFVGTKK36 pKa = 9.82SYY38 pKa = 11.22SGTTGFTDD46 pKa = 3.17VATLIGATYY55 pKa = 10.82DD56 pKa = 3.32SGTGKK61 pKa = 7.99MTQGLTKK68 pKa = 10.69AEE70 pKa = 3.97ISSMLFTKK78 pKa = 10.78SMGAYY83 pKa = 9.61NLNDD87 pKa = 3.49AVAWTYY93 pKa = 11.54DD94 pKa = 3.39YY95 pKa = 10.3GTVGDD100 pKa = 4.73GSDD103 pKa = 3.21ATVTDD108 pKa = 3.84GTTTVPVSILTSCGITGTSALADD131 pKa = 3.99GDD133 pKa = 4.34PVTGLTWEE141 pKa = 4.06MTNAEE146 pKa = 4.62VVTALQAAEE155 pKa = 4.33TNPDD159 pKa = 3.45IAWSYY164 pKa = 8.92STGSGYY170 pKa = 10.87VATHH174 pKa = 5.51TTTLGNSDD182 pKa = 4.9LNTGYY187 pKa = 10.89YY188 pKa = 9.59YY189 pKa = 10.96DD190 pKa = 3.7SAQGQFEE197 pKa = 4.24QGGTIVTAAEE207 pKa = 4.46AYY209 pKa = 9.04GIPADD214 pKa = 3.69TDD216 pKa = 3.57AVKK219 pKa = 9.37NTFTQNADD227 pKa = 3.03GSITLSTTTKK237 pKa = 8.17TKK239 pKa = 10.79DD240 pKa = 3.24YY241 pKa = 10.08IVQFNTSTGAFVSVNGEE258 pKa = 3.92DD259 pKa = 4.91DD260 pKa = 3.9VNLDD264 pKa = 3.76FAAGYY269 pKa = 11.46GNFKK273 pKa = 10.24DD274 pKa = 3.68INMDD278 pKa = 4.22FKK280 pKa = 10.93GTTMVDD286 pKa = 3.28NNGSSTLVMEE296 pKa = 4.84TGDD299 pKa = 3.81ADD301 pKa = 3.98GVGAGKK307 pKa = 10.68KK308 pKa = 9.21LGQMTGVSIQNNGMIYY324 pKa = 8.89GTYY327 pKa = 10.76DD328 pKa = 3.03NGNTKK333 pKa = 10.53LLGQIATASFANASGLLKK351 pKa = 10.55EE352 pKa = 4.55GEE354 pKa = 4.27NLYY357 pKa = 10.91SATLNSGEE365 pKa = 4.34FDD367 pKa = 4.86GIGQDD372 pKa = 3.46VTADD376 pKa = 3.65GGSMSSGVLEE386 pKa = 4.19MSNVDD391 pKa = 3.73LSAEE395 pKa = 4.03FTEE398 pKa = 5.13MITTQRR404 pKa = 11.84GFQANSRR411 pKa = 11.84IITTSDD417 pKa = 2.83SMLEE421 pKa = 3.66EE422 pKa = 5.03LINLKK427 pKa = 10.35RR428 pKa = 3.63
MM1 pKa = 7.61TNYY4 pKa = 10.26QSIFQTGTLEE14 pKa = 5.06SGFTYY19 pKa = 10.36NAPQVVDD26 pKa = 3.28NTSYY30 pKa = 11.53NFVGTKK36 pKa = 9.82SYY38 pKa = 11.22SGTTGFTDD46 pKa = 3.17VATLIGATYY55 pKa = 10.82DD56 pKa = 3.32SGTGKK61 pKa = 7.99MTQGLTKK68 pKa = 10.69AEE70 pKa = 3.97ISSMLFTKK78 pKa = 10.78SMGAYY83 pKa = 9.61NLNDD87 pKa = 3.49AVAWTYY93 pKa = 11.54DD94 pKa = 3.39YY95 pKa = 10.3GTVGDD100 pKa = 4.73GSDD103 pKa = 3.21ATVTDD108 pKa = 3.84GTTTVPVSILTSCGITGTSALADD131 pKa = 3.99GDD133 pKa = 4.34PVTGLTWEE141 pKa = 4.06MTNAEE146 pKa = 4.62VVTALQAAEE155 pKa = 4.33TNPDD159 pKa = 3.45IAWSYY164 pKa = 8.92STGSGYY170 pKa = 10.87VATHH174 pKa = 5.51TTTLGNSDD182 pKa = 4.9LNTGYY187 pKa = 10.89YY188 pKa = 9.59YY189 pKa = 10.96DD190 pKa = 3.7SAQGQFEE197 pKa = 4.24QGGTIVTAAEE207 pKa = 4.46AYY209 pKa = 9.04GIPADD214 pKa = 3.69TDD216 pKa = 3.57AVKK219 pKa = 9.37NTFTQNADD227 pKa = 3.03GSITLSTTTKK237 pKa = 8.17TKK239 pKa = 10.79DD240 pKa = 3.24YY241 pKa = 10.08IVQFNTSTGAFVSVNGEE258 pKa = 3.92DD259 pKa = 4.91DD260 pKa = 3.9VNLDD264 pKa = 3.76FAAGYY269 pKa = 11.46GNFKK273 pKa = 10.24DD274 pKa = 3.68INMDD278 pKa = 4.22FKK280 pKa = 10.93GTTMVDD286 pKa = 3.28NNGSSTLVMEE296 pKa = 4.84TGDD299 pKa = 3.81ADD301 pKa = 3.98GVGAGKK307 pKa = 10.68KK308 pKa = 9.21LGQMTGVSIQNNGMIYY324 pKa = 8.89GTYY327 pKa = 10.76DD328 pKa = 3.03NGNTKK333 pKa = 10.53LLGQIATASFANASGLLKK351 pKa = 10.55EE352 pKa = 4.55GEE354 pKa = 4.27NLYY357 pKa = 10.91SATLNSGEE365 pKa = 4.34FDD367 pKa = 4.86GIGQDD372 pKa = 3.46VTADD376 pKa = 3.65GGSMSSGVLEE386 pKa = 4.19MSNVDD391 pKa = 3.73LSAEE395 pKa = 4.03FTEE398 pKa = 5.13MITTQRR404 pKa = 11.84GFQANSRR411 pKa = 11.84IITTSDD417 pKa = 2.83SMLEE421 pKa = 3.66EE422 pKa = 5.03LINLKK427 pKa = 10.35RR428 pKa = 3.63
Molecular weight: 44.77 kDa
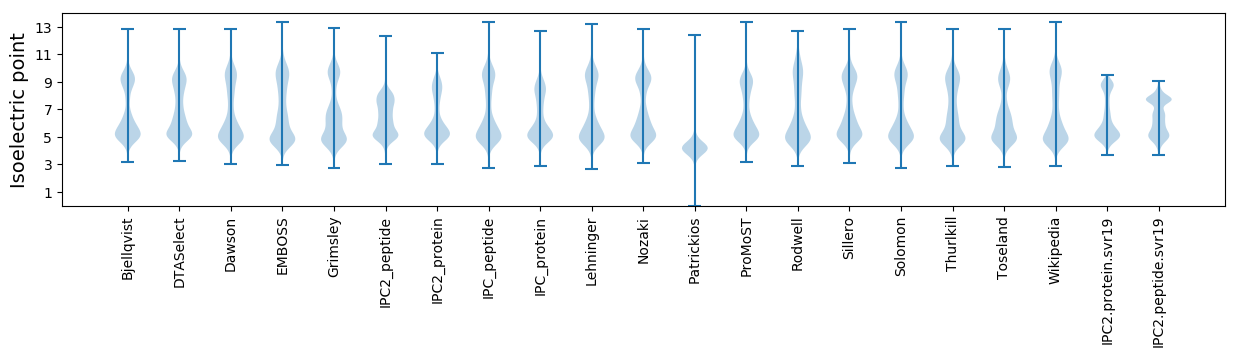
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A371AVF4|A0A371AVF4_9FIRM Uncharacterized protein OS=Anaerosacchriphilus polymeriproducens OX=1812858 GN=DWV06_09000 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.69VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.69VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1128028 |
24 |
4564 |
319.5 |
36.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.939 ± 0.044 | 1.355 ± 0.015 |
5.216 ± 0.03 | 7.578 ± 0.052 |
4.443 ± 0.037 | 6.448 ± 0.036 |
1.569 ± 0.016 | 9.17 ± 0.043 |
8.37 ± 0.034 | 8.986 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.851 ± 0.019 | 5.512 ± 0.037 |
2.833 ± 0.023 | 3.248 ± 0.027 |
3.493 ± 0.027 | 6.144 ± 0.029 |
5.184 ± 0.034 | 6.519 ± 0.032 |
0.826 ± 0.014 | 4.316 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
