
Mount Mabu Lophuromys virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Jeilongvirus; Lophuromys jeilongvirus 2
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
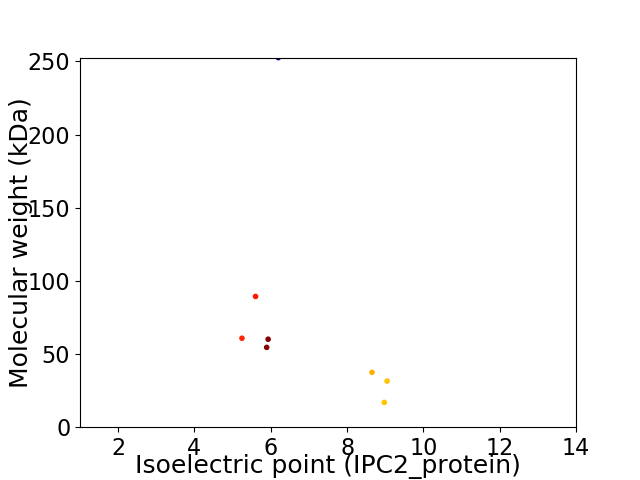
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
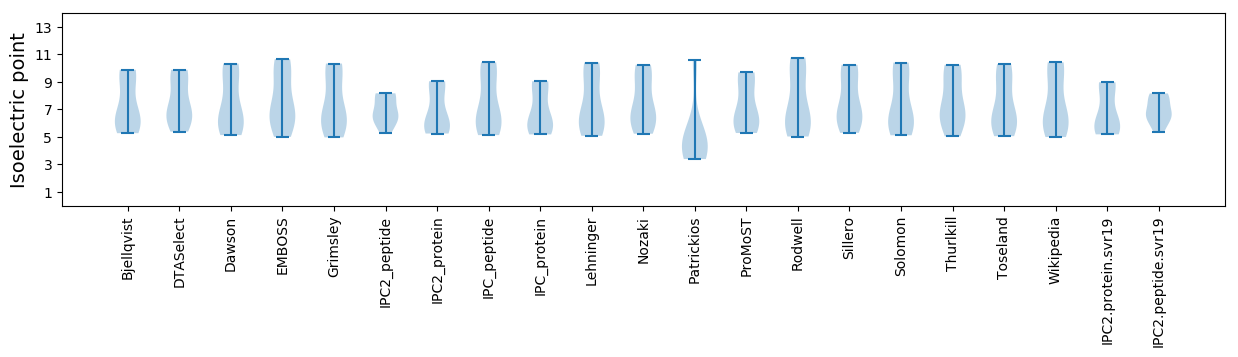
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1GJB4|A0A2P1GJB4_9MONO Putative TM protein OS=Mount Mabu Lophuromys virus 2 OX=2116560 GN=TM PE=4 SV=1
MM1 pKa = 7.42SRR3 pKa = 11.84LNSVLEE9 pKa = 3.93EE10 pKa = 3.68FRR12 pKa = 11.84EE13 pKa = 4.56FKK15 pKa = 10.95NNPPKK20 pKa = 10.51RR21 pKa = 11.84GLISSALHH29 pKa = 5.62GLKK32 pKa = 10.19KK33 pKa = 10.57SVIIPVPTFVDD44 pKa = 5.5PIQRR48 pKa = 11.84WYY50 pKa = 11.26FMVFMLEE57 pKa = 4.23LAWSNVASGSIMTGALLSLLSLFAEE82 pKa = 4.63NPGAMVRR89 pKa = 11.84SLLNDD94 pKa = 3.86PDD96 pKa = 5.16LDD98 pKa = 3.89VQIAEE103 pKa = 4.23VSRR106 pKa = 11.84ASKK109 pKa = 11.13DD110 pKa = 3.6EE111 pKa = 3.99IKK113 pKa = 10.8LATRR117 pKa = 11.84GKK119 pKa = 10.53NMEE122 pKa = 4.66DD123 pKa = 3.63YY124 pKa = 10.12EE125 pKa = 5.01KK126 pKa = 10.99EE127 pKa = 4.13LIRR130 pKa = 11.84IAGYY134 pKa = 10.52GPYY137 pKa = 9.71QGTKK141 pKa = 8.49TDD143 pKa = 3.89PFPFVKK149 pKa = 10.01KK150 pKa = 10.42DD151 pKa = 3.31QDD153 pKa = 3.34KK154 pKa = 10.56MITKK158 pKa = 7.73TTEE161 pKa = 3.77DD162 pKa = 3.41LQLAVQTVTIQIWILLTKK180 pKa = 10.49AVTATDD186 pKa = 3.53TAKK189 pKa = 10.56EE190 pKa = 4.22SEE192 pKa = 3.71NRR194 pKa = 11.84RR195 pKa = 11.84WVKK198 pKa = 10.28YY199 pKa = 8.73LQQRR203 pKa = 11.84RR204 pKa = 11.84ADD206 pKa = 3.9RR207 pKa = 11.84DD208 pKa = 3.7YY209 pKa = 11.53KK210 pKa = 11.38LEE212 pKa = 4.21DD213 pKa = 2.83TWLNLARR220 pKa = 11.84ARR222 pKa = 11.84IAEE225 pKa = 4.77DD226 pKa = 2.91ISVRR230 pKa = 11.84RR231 pKa = 11.84YY232 pKa = 7.94MVEE235 pKa = 4.0ILIEE239 pKa = 4.23TNKK242 pKa = 9.28MAGVKK247 pKa = 8.91TRR249 pKa = 11.84IVEE252 pKa = 4.32MISDD256 pKa = 3.73IGNYY260 pKa = 9.35ISEE263 pKa = 4.29AGMAGFFLTIKK274 pKa = 10.67YY275 pKa = 10.24GIEE278 pKa = 3.86TKK280 pKa = 10.96YY281 pKa = 10.24PALALNEE288 pKa = 4.12LQADD292 pKa = 4.15LSIILSLMKK301 pKa = 10.07TYY303 pKa = 10.66TSLGEE308 pKa = 3.87KK309 pKa = 9.73APYY312 pKa = 9.25MVILEE317 pKa = 4.66DD318 pKa = 5.19AIQTKK323 pKa = 9.64FSPGNYY329 pKa = 8.42PLLWSYY335 pKa = 12.02AMGVGSVLDD344 pKa = 3.37RR345 pKa = 11.84AVNNLNYY352 pKa = 8.54TRR354 pKa = 11.84PYY356 pKa = 10.0LEE358 pKa = 4.81HH359 pKa = 7.66SFFQLGEE366 pKa = 4.21TMVEE370 pKa = 4.02KK371 pKa = 10.78MEE373 pKa = 4.12GTVNNRR379 pKa = 11.84IAAEE383 pKa = 4.19LDD385 pKa = 3.43LSPEE389 pKa = 4.09QVQSIRR395 pKa = 11.84SIVKK399 pKa = 10.0LDD401 pKa = 3.47QATSGPRR408 pKa = 11.84VSGPQQKK415 pKa = 8.89TSGPRR420 pKa = 11.84PFEE423 pKa = 4.37LYY425 pKa = 10.27NAGDD429 pKa = 4.58IIPEE433 pKa = 3.98IEE435 pKa = 4.37EE436 pKa = 4.15EE437 pKa = 4.93VSDD440 pKa = 4.03EE441 pKa = 4.37DD442 pKa = 5.32KK443 pKa = 11.28KK444 pKa = 10.97DD445 pKa = 3.36TYY447 pKa = 11.16DD448 pKa = 3.18VSKK451 pKa = 10.96VVPVPNANSIFYY463 pKa = 10.93DD464 pKa = 4.06KK465 pKa = 11.22DD466 pKa = 3.4NQKK469 pKa = 11.06DD470 pKa = 3.62KK471 pKa = 11.47RR472 pKa = 11.84NLPKK476 pKa = 10.02PPKK479 pKa = 9.28PSRR482 pKa = 11.84SGKK485 pKa = 9.64KK486 pKa = 9.6VNNLKK491 pKa = 10.64EE492 pKa = 4.29SLNDD496 pKa = 3.6ALEE499 pKa = 4.37DD500 pKa = 4.0FVRR503 pKa = 11.84DD504 pKa = 3.16EE505 pKa = 6.06SMTQKK510 pKa = 10.62LNKK513 pKa = 9.57YY514 pKa = 10.38DD515 pKa = 4.2EE516 pKa = 5.63DD517 pKa = 4.08DD518 pKa = 3.46VSYY521 pKa = 10.04STDD524 pKa = 3.21QSPATMSDD532 pKa = 3.09LDD534 pKa = 3.81ALRR537 pKa = 11.84SS538 pKa = 3.64
MM1 pKa = 7.42SRR3 pKa = 11.84LNSVLEE9 pKa = 3.93EE10 pKa = 3.68FRR12 pKa = 11.84EE13 pKa = 4.56FKK15 pKa = 10.95NNPPKK20 pKa = 10.51RR21 pKa = 11.84GLISSALHH29 pKa = 5.62GLKK32 pKa = 10.19KK33 pKa = 10.57SVIIPVPTFVDD44 pKa = 5.5PIQRR48 pKa = 11.84WYY50 pKa = 11.26FMVFMLEE57 pKa = 4.23LAWSNVASGSIMTGALLSLLSLFAEE82 pKa = 4.63NPGAMVRR89 pKa = 11.84SLLNDD94 pKa = 3.86PDD96 pKa = 5.16LDD98 pKa = 3.89VQIAEE103 pKa = 4.23VSRR106 pKa = 11.84ASKK109 pKa = 11.13DD110 pKa = 3.6EE111 pKa = 3.99IKK113 pKa = 10.8LATRR117 pKa = 11.84GKK119 pKa = 10.53NMEE122 pKa = 4.66DD123 pKa = 3.63YY124 pKa = 10.12EE125 pKa = 5.01KK126 pKa = 10.99EE127 pKa = 4.13LIRR130 pKa = 11.84IAGYY134 pKa = 10.52GPYY137 pKa = 9.71QGTKK141 pKa = 8.49TDD143 pKa = 3.89PFPFVKK149 pKa = 10.01KK150 pKa = 10.42DD151 pKa = 3.31QDD153 pKa = 3.34KK154 pKa = 10.56MITKK158 pKa = 7.73TTEE161 pKa = 3.77DD162 pKa = 3.41LQLAVQTVTIQIWILLTKK180 pKa = 10.49AVTATDD186 pKa = 3.53TAKK189 pKa = 10.56EE190 pKa = 4.22SEE192 pKa = 3.71NRR194 pKa = 11.84RR195 pKa = 11.84WVKK198 pKa = 10.28YY199 pKa = 8.73LQQRR203 pKa = 11.84RR204 pKa = 11.84ADD206 pKa = 3.9RR207 pKa = 11.84DD208 pKa = 3.7YY209 pKa = 11.53KK210 pKa = 11.38LEE212 pKa = 4.21DD213 pKa = 2.83TWLNLARR220 pKa = 11.84ARR222 pKa = 11.84IAEE225 pKa = 4.77DD226 pKa = 2.91ISVRR230 pKa = 11.84RR231 pKa = 11.84YY232 pKa = 7.94MVEE235 pKa = 4.0ILIEE239 pKa = 4.23TNKK242 pKa = 9.28MAGVKK247 pKa = 8.91TRR249 pKa = 11.84IVEE252 pKa = 4.32MISDD256 pKa = 3.73IGNYY260 pKa = 9.35ISEE263 pKa = 4.29AGMAGFFLTIKK274 pKa = 10.67YY275 pKa = 10.24GIEE278 pKa = 3.86TKK280 pKa = 10.96YY281 pKa = 10.24PALALNEE288 pKa = 4.12LQADD292 pKa = 4.15LSIILSLMKK301 pKa = 10.07TYY303 pKa = 10.66TSLGEE308 pKa = 3.87KK309 pKa = 9.73APYY312 pKa = 9.25MVILEE317 pKa = 4.66DD318 pKa = 5.19AIQTKK323 pKa = 9.64FSPGNYY329 pKa = 8.42PLLWSYY335 pKa = 12.02AMGVGSVLDD344 pKa = 3.37RR345 pKa = 11.84AVNNLNYY352 pKa = 8.54TRR354 pKa = 11.84PYY356 pKa = 10.0LEE358 pKa = 4.81HH359 pKa = 7.66SFFQLGEE366 pKa = 4.21TMVEE370 pKa = 4.02KK371 pKa = 10.78MEE373 pKa = 4.12GTVNNRR379 pKa = 11.84IAAEE383 pKa = 4.19LDD385 pKa = 3.43LSPEE389 pKa = 4.09QVQSIRR395 pKa = 11.84SIVKK399 pKa = 10.0LDD401 pKa = 3.47QATSGPRR408 pKa = 11.84VSGPQQKK415 pKa = 8.89TSGPRR420 pKa = 11.84PFEE423 pKa = 4.37LYY425 pKa = 10.27NAGDD429 pKa = 4.58IIPEE433 pKa = 3.98IEE435 pKa = 4.37EE436 pKa = 4.15EE437 pKa = 4.93VSDD440 pKa = 4.03EE441 pKa = 4.37DD442 pKa = 5.32KK443 pKa = 11.28KK444 pKa = 10.97DD445 pKa = 3.36TYY447 pKa = 11.16DD448 pKa = 3.18VSKK451 pKa = 10.96VVPVPNANSIFYY463 pKa = 10.93DD464 pKa = 4.06KK465 pKa = 11.22DD466 pKa = 3.4NQKK469 pKa = 11.06DD470 pKa = 3.62KK471 pKa = 11.47RR472 pKa = 11.84NLPKK476 pKa = 10.02PPKK479 pKa = 9.28PSRR482 pKa = 11.84SGKK485 pKa = 9.64KK486 pKa = 9.6VNNLKK491 pKa = 10.64EE492 pKa = 4.29SLNDD496 pKa = 3.6ALEE499 pKa = 4.37DD500 pKa = 4.0FVRR503 pKa = 11.84DD504 pKa = 3.16EE505 pKa = 6.06SMTQKK510 pKa = 10.62LNKK513 pKa = 9.57YY514 pKa = 10.38DD515 pKa = 4.2EE516 pKa = 5.63DD517 pKa = 4.08DD518 pKa = 3.46VSYY521 pKa = 10.04STDD524 pKa = 3.21QSPATMSDD532 pKa = 3.09LDD534 pKa = 3.81ALRR537 pKa = 11.84SS538 pKa = 3.64
Molecular weight: 60.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1GJ92|A0A2P1GJ92_9MONO RNA-directed RNA polymerase L OS=Mount Mabu Lophuromys virus 2 OX=2116560 GN=L PE=3 SV=1
MM1 pKa = 7.51EE2 pKa = 5.31SALSTLSSRR11 pKa = 11.84IRR13 pKa = 11.84RR14 pKa = 11.84TFRR17 pKa = 11.84RR18 pKa = 11.84HH19 pKa = 4.34TGEE22 pKa = 3.99VQSINHH28 pKa = 5.52QPRR31 pKa = 11.84IEE33 pKa = 3.99QRR35 pKa = 11.84RR36 pKa = 11.84GRR38 pKa = 11.84PSQRR42 pKa = 11.84VRR44 pKa = 11.84VEE46 pKa = 4.02DD47 pKa = 3.88LSVNPGEE54 pKa = 4.01WEE56 pKa = 3.79AAKK59 pKa = 8.75EE60 pKa = 4.31TRR62 pKa = 11.84KK63 pKa = 8.67RR64 pKa = 11.84TAAEE68 pKa = 3.72VLEE71 pKa = 5.52LIDD74 pKa = 4.0HH75 pKa = 6.76LEE77 pKa = 4.25SKK79 pKa = 9.15GTEE82 pKa = 3.79VTAPRR87 pKa = 11.84TPRR90 pKa = 11.84EE91 pKa = 3.67QEE93 pKa = 4.08MITKK97 pKa = 10.49LGMLKK102 pKa = 9.94ILLNLVKK109 pKa = 9.34EE110 pKa = 4.5TGVIPDD116 pKa = 4.38PNKK119 pKa = 10.42QSIQQSGLLTRR130 pKa = 11.84VEE132 pKa = 4.14MEE134 pKa = 4.47NLGEE138 pKa = 4.41MIPALKK144 pKa = 10.5SILKK148 pKa = 9.84NN149 pKa = 3.45
MM1 pKa = 7.51EE2 pKa = 5.31SALSTLSSRR11 pKa = 11.84IRR13 pKa = 11.84RR14 pKa = 11.84TFRR17 pKa = 11.84RR18 pKa = 11.84HH19 pKa = 4.34TGEE22 pKa = 3.99VQSINHH28 pKa = 5.52QPRR31 pKa = 11.84IEE33 pKa = 3.99QRR35 pKa = 11.84RR36 pKa = 11.84GRR38 pKa = 11.84PSQRR42 pKa = 11.84VRR44 pKa = 11.84VEE46 pKa = 4.02DD47 pKa = 3.88LSVNPGEE54 pKa = 4.01WEE56 pKa = 3.79AAKK59 pKa = 8.75EE60 pKa = 4.31TRR62 pKa = 11.84KK63 pKa = 8.67RR64 pKa = 11.84TAAEE68 pKa = 3.72VLEE71 pKa = 5.52LIDD74 pKa = 4.0HH75 pKa = 6.76LEE77 pKa = 4.25SKK79 pKa = 9.15GTEE82 pKa = 3.79VTAPRR87 pKa = 11.84TPRR90 pKa = 11.84EE91 pKa = 3.67QEE93 pKa = 4.08MITKK97 pKa = 10.49LGMLKK102 pKa = 9.94ILLNLVKK109 pKa = 9.34EE110 pKa = 4.5TGVIPDD116 pKa = 4.38PNKK119 pKa = 10.42QSIQQSGLLTRR130 pKa = 11.84VEE132 pKa = 4.14MEE134 pKa = 4.47NLGEE138 pKa = 4.41MIPALKK144 pKa = 10.5SILKK148 pKa = 9.84NN149 pKa = 3.45
Molecular weight: 16.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5346 |
149 |
2188 |
668.3 |
75.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.863 ± 0.361 | 1.553 ± 0.373 |
5.724 ± 0.414 | 5.556 ± 0.499 |
3.236 ± 0.351 | 5.743 ± 0.709 |
2.132 ± 0.411 | 7.838 ± 0.535 |
6.023 ± 0.675 | 9.39 ± 0.814 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.768 ± 0.186 | 6.192 ± 0.389 |
3.984 ± 0.297 | 3.498 ± 0.287 |
5.051 ± 0.336 | 8.474 ± 0.395 |
6.921 ± 1.003 | 5.855 ± 0.529 |
1.066 ± 0.141 | 4.134 ± 0.56 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
