
Alteromonas lipolytica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Alteromonas
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
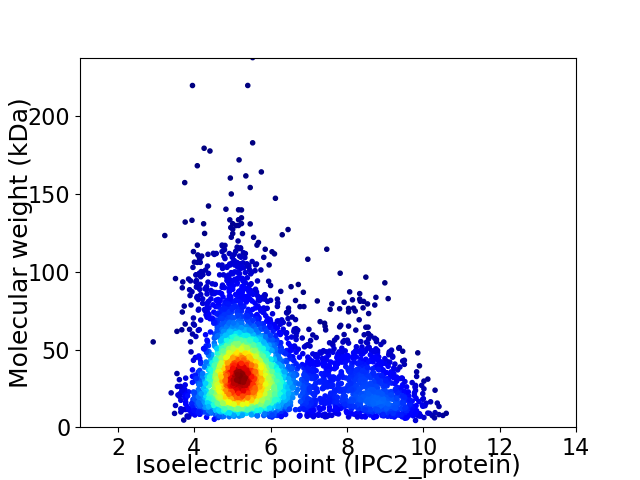
Virtual 2D-PAGE plot for 4316 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E8FGX6|A0A1E8FGX6_9ALTE Lytic transglycosylase OS=Alteromonas lipolytica OX=1856405 GN=BFC17_14120 PE=4 SV=1
MM1 pKa = 7.71KK2 pKa = 10.48LKK4 pKa = 10.36QLASAVILATSGVAIAPLATAATYY28 pKa = 10.57DD29 pKa = 3.69LSLVPVQDD37 pKa = 3.59KK38 pKa = 11.11VFLNFAQSIDD48 pKa = 3.58NTGTMVVTAQGGYY61 pKa = 9.92NPPIDD66 pKa = 5.18LSLLDD71 pKa = 5.2FEE73 pKa = 5.94DD74 pKa = 3.42EE75 pKa = 4.15TFIARR80 pKa = 11.84FADD83 pKa = 3.91PEE85 pKa = 4.1AVEE88 pKa = 3.98QGIFSSADD96 pKa = 3.36YY97 pKa = 10.91QEE99 pKa = 4.71LYY101 pKa = 10.8AYY103 pKa = 9.4IVNARR108 pKa = 11.84GAGNTTFQPIASYY121 pKa = 10.76RR122 pKa = 11.84SYY124 pKa = 11.49KK125 pKa = 10.31SDD127 pKa = 3.02TVTADD132 pKa = 4.7LIPGFDD138 pKa = 4.15AFNDD142 pKa = 4.57AINDD146 pKa = 3.55YY147 pKa = 9.51WRR149 pKa = 11.84QNYY152 pKa = 8.13TSVYY156 pKa = 9.91DD157 pKa = 3.99SVGGDD162 pKa = 3.62YY163 pKa = 10.83FVGIANAPFFTVDD176 pKa = 3.26YY177 pKa = 10.0TNDD180 pKa = 3.67DD181 pKa = 3.71GDD183 pKa = 3.75EE184 pKa = 3.89FTYY187 pKa = 10.27IINNAQTQGFVEE199 pKa = 4.6VNGNATPLPSALADD213 pKa = 3.67APIPGYY219 pKa = 9.44SEE221 pKa = 3.91PFAINQSLQVVGWASVDD238 pKa = 3.5VTDD241 pKa = 6.18DD242 pKa = 3.61FQEE245 pKa = 5.84DD246 pKa = 3.07IDD248 pKa = 4.37ACLDD252 pKa = 3.52DD253 pKa = 4.44EE254 pKa = 4.94TRR256 pKa = 11.84GSQPFDD262 pKa = 3.11VCLYY266 pKa = 10.12NLSLNVTTGTYY277 pKa = 9.61RR278 pKa = 11.84RR279 pKa = 11.84AAIWQLDD286 pKa = 3.51AQGNVLSTTPYY297 pKa = 10.97GMAFEE302 pKa = 5.09PDD304 pKa = 2.92EE305 pKa = 4.63DD306 pKa = 4.53EE307 pKa = 4.56YY308 pKa = 10.73VTGYY312 pKa = 10.51VSSARR317 pKa = 11.84DD318 pKa = 3.37INDD321 pKa = 2.96NGIAVGYY328 pKa = 8.64AHH330 pKa = 6.85NGEE333 pKa = 4.37FVLYY337 pKa = 8.08TLPSAQPRR345 pKa = 11.84RR346 pKa = 11.84FTFPQEE352 pKa = 3.86VATTFVDD359 pKa = 4.69GVVTEE364 pKa = 4.01ILPRR368 pKa = 11.84EE369 pKa = 4.29DD370 pKa = 3.77SLSSYY375 pKa = 10.55AQGINNDD382 pKa = 3.26NWVMGTVLEE391 pKa = 4.54EE392 pKa = 4.13NNGFARR398 pKa = 11.84QHH400 pKa = 6.08LFVHH404 pKa = 6.15NLDD407 pKa = 3.63SGEE410 pKa = 4.59SIFPQGFFQTAGVRR424 pKa = 11.84PRR426 pKa = 11.84AINNNGIVVGEE437 pKa = 4.31GEE439 pKa = 4.43YY440 pKa = 10.9EE441 pKa = 4.11SDD443 pKa = 3.26VQYY446 pKa = 11.32DD447 pKa = 3.64RR448 pKa = 11.84QVRR451 pKa = 11.84AFMYY455 pKa = 9.92DD456 pKa = 2.92INVGDD461 pKa = 5.22FIDD464 pKa = 5.26LNTLLSCEE472 pKa = 3.74QRR474 pKa = 11.84EE475 pKa = 4.3QYY477 pKa = 10.91RR478 pKa = 11.84LVSAADD484 pKa = 3.3INDD487 pKa = 3.8DD488 pKa = 4.2NEE490 pKa = 4.71IIANALYY497 pKa = 10.7LDD499 pKa = 3.67TQRR502 pKa = 11.84YY503 pKa = 8.14IDD505 pKa = 4.03GEE507 pKa = 4.47EE508 pKa = 4.07VLTTAGDD515 pKa = 3.76TIDD518 pKa = 3.46ANRR521 pKa = 11.84IVAVKK526 pKa = 8.13LTPNGTGSVEE536 pKa = 3.93QCDD539 pKa = 3.32AAEE542 pKa = 4.01EE543 pKa = 4.0QNYY546 pKa = 7.9EE547 pKa = 4.12RR548 pKa = 11.84KK549 pKa = 10.04GATGTLYY556 pKa = 11.24GLLTLGLFAFLRR568 pKa = 11.84RR569 pKa = 11.84LSFRR573 pKa = 3.77
MM1 pKa = 7.71KK2 pKa = 10.48LKK4 pKa = 10.36QLASAVILATSGVAIAPLATAATYY28 pKa = 10.57DD29 pKa = 3.69LSLVPVQDD37 pKa = 3.59KK38 pKa = 11.11VFLNFAQSIDD48 pKa = 3.58NTGTMVVTAQGGYY61 pKa = 9.92NPPIDD66 pKa = 5.18LSLLDD71 pKa = 5.2FEE73 pKa = 5.94DD74 pKa = 3.42EE75 pKa = 4.15TFIARR80 pKa = 11.84FADD83 pKa = 3.91PEE85 pKa = 4.1AVEE88 pKa = 3.98QGIFSSADD96 pKa = 3.36YY97 pKa = 10.91QEE99 pKa = 4.71LYY101 pKa = 10.8AYY103 pKa = 9.4IVNARR108 pKa = 11.84GAGNTTFQPIASYY121 pKa = 10.76RR122 pKa = 11.84SYY124 pKa = 11.49KK125 pKa = 10.31SDD127 pKa = 3.02TVTADD132 pKa = 4.7LIPGFDD138 pKa = 4.15AFNDD142 pKa = 4.57AINDD146 pKa = 3.55YY147 pKa = 9.51WRR149 pKa = 11.84QNYY152 pKa = 8.13TSVYY156 pKa = 9.91DD157 pKa = 3.99SVGGDD162 pKa = 3.62YY163 pKa = 10.83FVGIANAPFFTVDD176 pKa = 3.26YY177 pKa = 10.0TNDD180 pKa = 3.67DD181 pKa = 3.71GDD183 pKa = 3.75EE184 pKa = 3.89FTYY187 pKa = 10.27IINNAQTQGFVEE199 pKa = 4.6VNGNATPLPSALADD213 pKa = 3.67APIPGYY219 pKa = 9.44SEE221 pKa = 3.91PFAINQSLQVVGWASVDD238 pKa = 3.5VTDD241 pKa = 6.18DD242 pKa = 3.61FQEE245 pKa = 5.84DD246 pKa = 3.07IDD248 pKa = 4.37ACLDD252 pKa = 3.52DD253 pKa = 4.44EE254 pKa = 4.94TRR256 pKa = 11.84GSQPFDD262 pKa = 3.11VCLYY266 pKa = 10.12NLSLNVTTGTYY277 pKa = 9.61RR278 pKa = 11.84RR279 pKa = 11.84AAIWQLDD286 pKa = 3.51AQGNVLSTTPYY297 pKa = 10.97GMAFEE302 pKa = 5.09PDD304 pKa = 2.92EE305 pKa = 4.63DD306 pKa = 4.53EE307 pKa = 4.56YY308 pKa = 10.73VTGYY312 pKa = 10.51VSSARR317 pKa = 11.84DD318 pKa = 3.37INDD321 pKa = 2.96NGIAVGYY328 pKa = 8.64AHH330 pKa = 6.85NGEE333 pKa = 4.37FVLYY337 pKa = 8.08TLPSAQPRR345 pKa = 11.84RR346 pKa = 11.84FTFPQEE352 pKa = 3.86VATTFVDD359 pKa = 4.69GVVTEE364 pKa = 4.01ILPRR368 pKa = 11.84EE369 pKa = 4.29DD370 pKa = 3.77SLSSYY375 pKa = 10.55AQGINNDD382 pKa = 3.26NWVMGTVLEE391 pKa = 4.54EE392 pKa = 4.13NNGFARR398 pKa = 11.84QHH400 pKa = 6.08LFVHH404 pKa = 6.15NLDD407 pKa = 3.63SGEE410 pKa = 4.59SIFPQGFFQTAGVRR424 pKa = 11.84PRR426 pKa = 11.84AINNNGIVVGEE437 pKa = 4.31GEE439 pKa = 4.43YY440 pKa = 10.9EE441 pKa = 4.11SDD443 pKa = 3.26VQYY446 pKa = 11.32DD447 pKa = 3.64RR448 pKa = 11.84QVRR451 pKa = 11.84AFMYY455 pKa = 9.92DD456 pKa = 2.92INVGDD461 pKa = 5.22FIDD464 pKa = 5.26LNTLLSCEE472 pKa = 3.74QRR474 pKa = 11.84EE475 pKa = 4.3QYY477 pKa = 10.91RR478 pKa = 11.84LVSAADD484 pKa = 3.3INDD487 pKa = 3.8DD488 pKa = 4.2NEE490 pKa = 4.71IIANALYY497 pKa = 10.7LDD499 pKa = 3.67TQRR502 pKa = 11.84YY503 pKa = 8.14IDD505 pKa = 4.03GEE507 pKa = 4.47EE508 pKa = 4.07VLTTAGDD515 pKa = 3.76TIDD518 pKa = 3.46ANRR521 pKa = 11.84IVAVKK526 pKa = 8.13LTPNGTGSVEE536 pKa = 3.93QCDD539 pKa = 3.32AAEE542 pKa = 4.01EE543 pKa = 4.0QNYY546 pKa = 7.9EE547 pKa = 4.12RR548 pKa = 11.84KK549 pKa = 10.04GATGTLYY556 pKa = 11.24GLLTLGLFAFLRR568 pKa = 11.84RR569 pKa = 11.84LSFRR573 pKa = 3.77
Molecular weight: 62.95 kDa
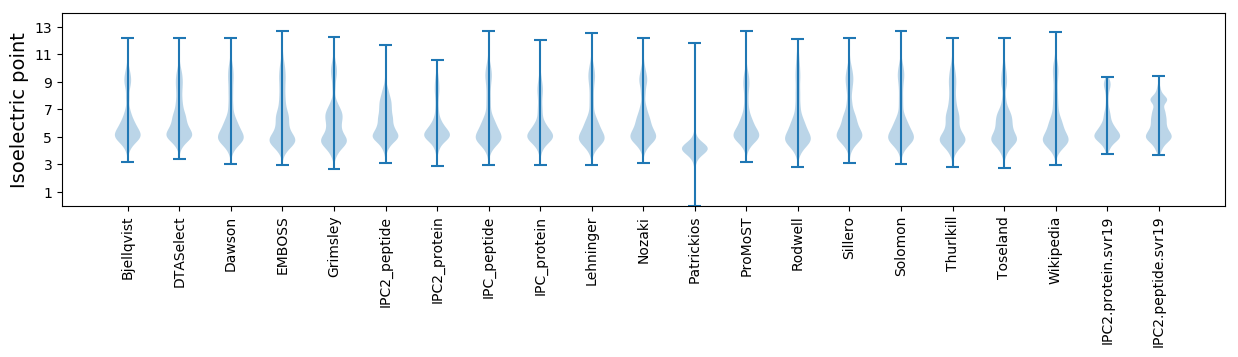
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E8FG89|A0A1E8FG89_9ALTE Sigma-54-dependent Fis family transcriptional regulator OS=Alteromonas lipolytica OX=1856405 GN=BFC17_13510 PE=4 SV=1
MM1 pKa = 7.82ASTIVLPPATVSRR14 pKa = 11.84IIEE17 pKa = 4.3MAWEE21 pKa = 4.07DD22 pKa = 3.71RR23 pKa = 11.84TPFEE27 pKa = 4.98AIEE30 pKa = 4.07RR31 pKa = 11.84EE32 pKa = 4.2YY33 pKa = 11.41GLSQDD38 pKa = 3.34GVIRR42 pKa = 11.84LMRR45 pKa = 11.84RR46 pKa = 11.84EE47 pKa = 4.36LKK49 pKa = 10.4PSSFRR54 pKa = 11.84LWRR57 pKa = 11.84KK58 pKa = 7.14RR59 pKa = 11.84TQGRR63 pKa = 11.84TTKK66 pKa = 10.04HH67 pKa = 5.59LKK69 pKa = 10.05LRR71 pKa = 11.84SPDD74 pKa = 2.99ISRR77 pKa = 11.84GYY79 pKa = 10.85CPTQYY84 pKa = 10.69KK85 pKa = 9.43QRR87 pKa = 3.81
MM1 pKa = 7.82ASTIVLPPATVSRR14 pKa = 11.84IIEE17 pKa = 4.3MAWEE21 pKa = 4.07DD22 pKa = 3.71RR23 pKa = 11.84TPFEE27 pKa = 4.98AIEE30 pKa = 4.07RR31 pKa = 11.84EE32 pKa = 4.2YY33 pKa = 11.41GLSQDD38 pKa = 3.34GVIRR42 pKa = 11.84LMRR45 pKa = 11.84RR46 pKa = 11.84EE47 pKa = 4.36LKK49 pKa = 10.4PSSFRR54 pKa = 11.84LWRR57 pKa = 11.84KK58 pKa = 7.14RR59 pKa = 11.84TQGRR63 pKa = 11.84TTKK66 pKa = 10.04HH67 pKa = 5.59LKK69 pKa = 10.05LRR71 pKa = 11.84SPDD74 pKa = 2.99ISRR77 pKa = 11.84GYY79 pKa = 10.85CPTQYY84 pKa = 10.69KK85 pKa = 9.43QRR87 pKa = 3.81
Molecular weight: 10.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1473345 |
38 |
2096 |
341.4 |
37.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.577 ± 0.034 | 1.005 ± 0.013 |
5.683 ± 0.032 | 6.029 ± 0.036 |
4.133 ± 0.025 | 6.957 ± 0.036 |
2.16 ± 0.018 | 5.95 ± 0.026 |
4.567 ± 0.032 | 10.321 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.512 ± 0.017 | 4.096 ± 0.026 |
4.154 ± 0.022 | 4.69 ± 0.032 |
4.679 ± 0.029 | 6.45 ± 0.026 |
5.586 ± 0.031 | 7.018 ± 0.028 |
1.277 ± 0.016 | 3.155 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
