
Alces alces faeces associated genomovirus MP84
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus alces1
Average proteome isoelectric point is 7.43
Get precalculated fractions of proteins
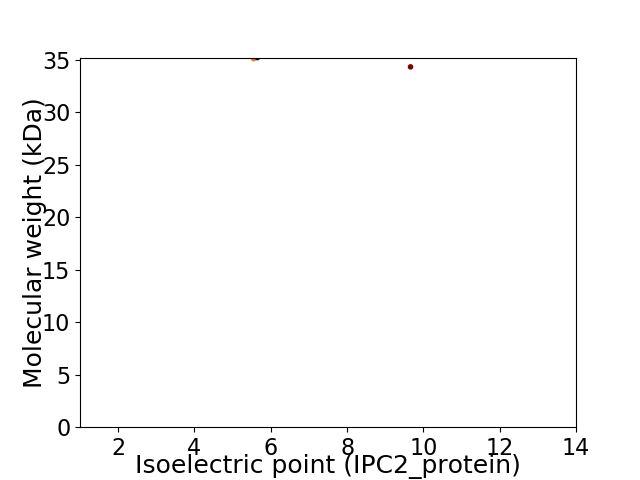
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CK60|A0A2Z5CK60_9VIRU Replication-associated protein OS=Alces alces faeces associated genomovirus MP84 OX=2219116 PE=3 SV=1
MM1 pKa = 7.88PRR3 pKa = 11.84FLCNASHH10 pKa = 7.43FLITYY15 pKa = 7.23AQCGDD20 pKa = 3.42LSEE23 pKa = 4.27WRR25 pKa = 11.84VLDD28 pKa = 4.01CFTLLGAQCIIARR41 pKa = 11.84EE42 pKa = 3.97YY43 pKa = 11.19HH44 pKa = 6.63EE45 pKa = 5.57DD46 pKa = 3.53LGIHH50 pKa = 5.87LHH52 pKa = 5.56VFVDD56 pKa = 4.71FRR58 pKa = 11.84RR59 pKa = 11.84KK60 pKa = 8.85FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84VDD67 pKa = 2.84IFDD70 pKa = 3.17VDD72 pKa = 3.57GRR74 pKa = 11.84HH75 pKa = 6.05PNIKK79 pKa = 9.98RR80 pKa = 11.84SWGTPEE86 pKa = 4.27KK87 pKa = 10.76GYY89 pKa = 10.97DD90 pKa = 3.47YY91 pKa = 10.69AIKK94 pKa = 10.87DD95 pKa = 3.59GDD97 pKa = 4.01VVAGGLEE104 pKa = 4.18RR105 pKa = 11.84PDD107 pKa = 3.36GTFGSGKK114 pKa = 9.79NFDD117 pKa = 4.27LWSQIAGAGSKK128 pKa = 11.05AEE130 pKa = 4.04FWDD133 pKa = 3.76LCEE136 pKa = 4.22EE137 pKa = 4.73LDD139 pKa = 4.84PKK141 pKa = 11.48SMWCSFGQLQKK152 pKa = 10.65FADD155 pKa = 3.31WRR157 pKa = 11.84FAEE160 pKa = 4.67VPTEE164 pKa = 3.85YY165 pKa = 10.89ANPGGFEE172 pKa = 4.08FVDD175 pKa = 4.28GNDD178 pKa = 4.25DD179 pKa = 3.56GRR181 pKa = 11.84TAPMSLVLYY190 pKa = 10.49GEE192 pKa = 4.26SRR194 pKa = 11.84TGKK197 pKa = 7.35TLWARR202 pKa = 11.84SLGPHH207 pKa = 7.22IYY209 pKa = 10.46NVGLVSGEE217 pKa = 4.0EE218 pKa = 4.6CSRR221 pKa = 11.84APHH224 pKa = 5.21VKK226 pKa = 10.21YY227 pKa = 10.68AVFDD231 pKa = 4.9DD232 pKa = 3.28IRR234 pKa = 11.84GGIKK238 pKa = 9.97FFPAFKK244 pKa = 9.79EE245 pKa = 4.03WLGGQSTVCVKK256 pKa = 10.49RR257 pKa = 11.84LYY259 pKa = 10.38RR260 pKa = 11.84DD261 pKa = 3.5PKK263 pKa = 10.22LVTWGKK269 pKa = 9.15PSIWVSNDD277 pKa = 3.26DD278 pKa = 4.22PRR280 pKa = 11.84HH281 pKa = 5.73AMDD284 pKa = 5.22PSDD287 pKa = 4.5VSWLEE292 pKa = 3.46ANARR296 pKa = 11.84FIEE299 pKa = 3.97ITEE302 pKa = 4.67PIFRR306 pKa = 11.84ANTGG310 pKa = 3.1
MM1 pKa = 7.88PRR3 pKa = 11.84FLCNASHH10 pKa = 7.43FLITYY15 pKa = 7.23AQCGDD20 pKa = 3.42LSEE23 pKa = 4.27WRR25 pKa = 11.84VLDD28 pKa = 4.01CFTLLGAQCIIARR41 pKa = 11.84EE42 pKa = 3.97YY43 pKa = 11.19HH44 pKa = 6.63EE45 pKa = 5.57DD46 pKa = 3.53LGIHH50 pKa = 5.87LHH52 pKa = 5.56VFVDD56 pKa = 4.71FRR58 pKa = 11.84RR59 pKa = 11.84KK60 pKa = 8.85FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84VDD67 pKa = 2.84IFDD70 pKa = 3.17VDD72 pKa = 3.57GRR74 pKa = 11.84HH75 pKa = 6.05PNIKK79 pKa = 9.98RR80 pKa = 11.84SWGTPEE86 pKa = 4.27KK87 pKa = 10.76GYY89 pKa = 10.97DD90 pKa = 3.47YY91 pKa = 10.69AIKK94 pKa = 10.87DD95 pKa = 3.59GDD97 pKa = 4.01VVAGGLEE104 pKa = 4.18RR105 pKa = 11.84PDD107 pKa = 3.36GTFGSGKK114 pKa = 9.79NFDD117 pKa = 4.27LWSQIAGAGSKK128 pKa = 11.05AEE130 pKa = 4.04FWDD133 pKa = 3.76LCEE136 pKa = 4.22EE137 pKa = 4.73LDD139 pKa = 4.84PKK141 pKa = 11.48SMWCSFGQLQKK152 pKa = 10.65FADD155 pKa = 3.31WRR157 pKa = 11.84FAEE160 pKa = 4.67VPTEE164 pKa = 3.85YY165 pKa = 10.89ANPGGFEE172 pKa = 4.08FVDD175 pKa = 4.28GNDD178 pKa = 4.25DD179 pKa = 3.56GRR181 pKa = 11.84TAPMSLVLYY190 pKa = 10.49GEE192 pKa = 4.26SRR194 pKa = 11.84TGKK197 pKa = 7.35TLWARR202 pKa = 11.84SLGPHH207 pKa = 7.22IYY209 pKa = 10.46NVGLVSGEE217 pKa = 4.0EE218 pKa = 4.6CSRR221 pKa = 11.84APHH224 pKa = 5.21VKK226 pKa = 10.21YY227 pKa = 10.68AVFDD231 pKa = 4.9DD232 pKa = 3.28IRR234 pKa = 11.84GGIKK238 pKa = 9.97FFPAFKK244 pKa = 9.79EE245 pKa = 4.03WLGGQSTVCVKK256 pKa = 10.49RR257 pKa = 11.84LYY259 pKa = 10.38RR260 pKa = 11.84DD261 pKa = 3.5PKK263 pKa = 10.22LVTWGKK269 pKa = 9.15PSIWVSNDD277 pKa = 3.26DD278 pKa = 4.22PRR280 pKa = 11.84HH281 pKa = 5.73AMDD284 pKa = 5.22PSDD287 pKa = 4.5VSWLEE292 pKa = 3.46ANARR296 pKa = 11.84FIEE299 pKa = 3.97ITEE302 pKa = 4.67PIFRR306 pKa = 11.84ANTGG310 pKa = 3.1
Molecular weight: 35.12 kDa
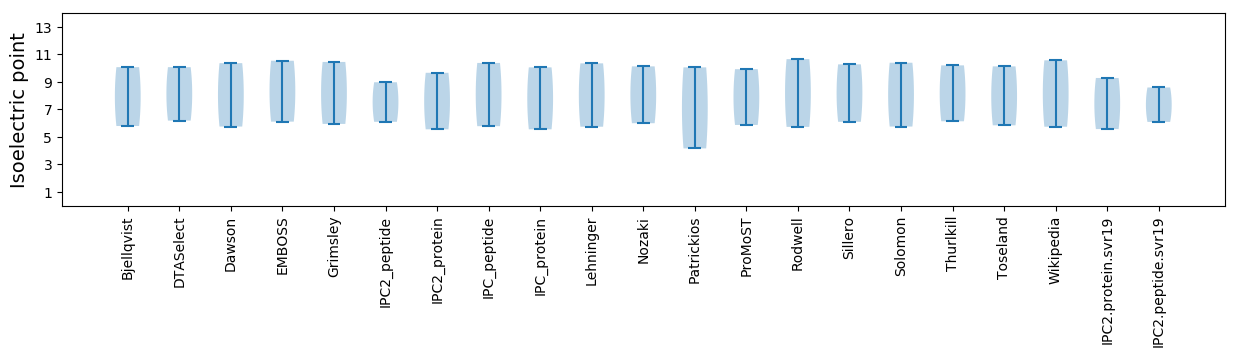
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CK60|A0A2Z5CK60_9VIRU Replication-associated protein OS=Alces alces faeces associated genomovirus MP84 OX=2219116 PE=3 SV=1
MM1 pKa = 8.1AYY3 pKa = 10.29RR4 pKa = 11.84STKK7 pKa = 9.25RR8 pKa = 11.84RR9 pKa = 11.84SPVRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 8.77PARR19 pKa = 11.84KK20 pKa = 8.78YY21 pKa = 10.63GGRR24 pKa = 11.84TTRR27 pKa = 11.84RR28 pKa = 11.84SYY30 pKa = 11.03AKK32 pKa = 10.03KK33 pKa = 8.36RR34 pKa = 11.84TYY36 pKa = 10.13RR37 pKa = 11.84KK38 pKa = 9.41KK39 pKa = 10.78ASSKK43 pKa = 9.65MSNKK47 pKa = 10.07RR48 pKa = 11.84ILNLTSIKK56 pKa = 10.5KK57 pKa = 9.84RR58 pKa = 11.84DD59 pKa = 3.76TMLSAYY65 pKa = 10.05QLGSTVNPQPGAYY78 pKa = 8.42PVQGTGGGNGGNTTVLLWCATARR101 pKa = 11.84DD102 pKa = 3.84NTVISGGGSGNRR114 pKa = 11.84FTPATRR120 pKa = 11.84TATTCFMRR128 pKa = 11.84GLAEE132 pKa = 4.03NVEE135 pKa = 4.47LSTNTGVAWQWRR147 pKa = 11.84RR148 pKa = 11.84ICFTFKK154 pKa = 11.11DD155 pKa = 4.2PGALSQTQTYY165 pKa = 9.9FLEE168 pKa = 4.71TSSGFQRR175 pKa = 11.84NNYY178 pKa = 9.01NVTSGSSPDD187 pKa = 3.44STNLEE192 pKa = 4.09NLRR195 pKa = 11.84QLVFRR200 pKa = 11.84GQLGTDD206 pKa = 2.82WSSYY210 pKa = 7.89LTAPVDD216 pKa = 3.92NSRR219 pKa = 11.84CTIKK223 pKa = 10.57YY224 pKa = 10.32DD225 pKa = 2.84KK226 pKa = 8.17TTNVNSGNANGKK238 pKa = 9.0LVNRR242 pKa = 11.84KK243 pKa = 8.13LWMPMNSNLVYY254 pKa = 10.84DD255 pKa = 4.53DD256 pKa = 5.43DD257 pKa = 4.34EE258 pKa = 6.52LGGAEE263 pKa = 4.05AARR266 pKa = 11.84PYY268 pKa = 10.82SIGGKK273 pKa = 9.45QGMGDD278 pKa = 4.24YY279 pKa = 10.98YY280 pKa = 11.77VMDD283 pKa = 3.99IFQAGVGATSSDD295 pKa = 3.62LLSFLPQATLYY306 pKa = 9.14WHH308 pKa = 6.98EE309 pKa = 4.28KK310 pKa = 9.3
MM1 pKa = 8.1AYY3 pKa = 10.29RR4 pKa = 11.84STKK7 pKa = 9.25RR8 pKa = 11.84RR9 pKa = 11.84SPVRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 8.77PARR19 pKa = 11.84KK20 pKa = 8.78YY21 pKa = 10.63GGRR24 pKa = 11.84TTRR27 pKa = 11.84RR28 pKa = 11.84SYY30 pKa = 11.03AKK32 pKa = 10.03KK33 pKa = 8.36RR34 pKa = 11.84TYY36 pKa = 10.13RR37 pKa = 11.84KK38 pKa = 9.41KK39 pKa = 10.78ASSKK43 pKa = 9.65MSNKK47 pKa = 10.07RR48 pKa = 11.84ILNLTSIKK56 pKa = 10.5KK57 pKa = 9.84RR58 pKa = 11.84DD59 pKa = 3.76TMLSAYY65 pKa = 10.05QLGSTVNPQPGAYY78 pKa = 8.42PVQGTGGGNGGNTTVLLWCATARR101 pKa = 11.84DD102 pKa = 3.84NTVISGGGSGNRR114 pKa = 11.84FTPATRR120 pKa = 11.84TATTCFMRR128 pKa = 11.84GLAEE132 pKa = 4.03NVEE135 pKa = 4.47LSTNTGVAWQWRR147 pKa = 11.84RR148 pKa = 11.84ICFTFKK154 pKa = 11.11DD155 pKa = 4.2PGALSQTQTYY165 pKa = 9.9FLEE168 pKa = 4.71TSSGFQRR175 pKa = 11.84NNYY178 pKa = 9.01NVTSGSSPDD187 pKa = 3.44STNLEE192 pKa = 4.09NLRR195 pKa = 11.84QLVFRR200 pKa = 11.84GQLGTDD206 pKa = 2.82WSSYY210 pKa = 7.89LTAPVDD216 pKa = 3.92NSRR219 pKa = 11.84CTIKK223 pKa = 10.57YY224 pKa = 10.32DD225 pKa = 2.84KK226 pKa = 8.17TTNVNSGNANGKK238 pKa = 9.0LVNRR242 pKa = 11.84KK243 pKa = 8.13LWMPMNSNLVYY254 pKa = 10.84DD255 pKa = 4.53DD256 pKa = 5.43DD257 pKa = 4.34EE258 pKa = 6.52LGGAEE263 pKa = 4.05AARR266 pKa = 11.84PYY268 pKa = 10.82SIGGKK273 pKa = 9.45QGMGDD278 pKa = 4.24YY279 pKa = 10.98YY280 pKa = 11.77VMDD283 pKa = 3.99IFQAGVGATSSDD295 pKa = 3.62LLSFLPQATLYY306 pKa = 9.14WHH308 pKa = 6.98EE309 pKa = 4.28KK310 pKa = 9.3
Molecular weight: 34.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
620 |
310 |
310 |
310.0 |
34.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.774 ± 0.0 | 1.935 ± 0.482 |
6.129 ± 1.446 | 4.032 ± 1.326 |
4.839 ± 1.446 | 9.839 ± 0.121 |
1.452 ± 0.844 | 3.548 ± 0.964 |
5.161 ± 0.241 | 7.258 ± 0.121 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.935 ± 0.482 | 5.0 ± 1.567 |
4.516 ± 0.482 | 2.903 ± 0.723 |
7.742 ± 0.482 | 7.581 ± 1.085 |
7.097 ± 2.411 | 5.484 ± 0.482 |
2.742 ± 0.603 | 4.032 ± 0.844 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
