
Firmicutes bacterium CAG:791
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
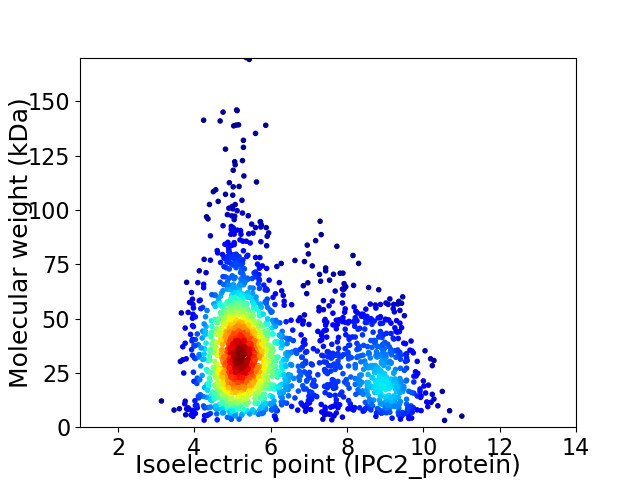
Virtual 2D-PAGE plot for 2175 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5C9X6|R5C9X6_9FIRM DNA replication and repair protein RecF OS=Firmicutes bacterium CAG:791 OX=1262993 GN=recF PE=3 SV=1
MM1 pKa = 7.81KK2 pKa = 10.32KK3 pKa = 10.3RR4 pKa = 11.84IVSVLMAGALASAMLAGCGNSSAPAASAAAEE35 pKa = 4.18DD36 pKa = 4.01TAEE39 pKa = 4.36DD40 pKa = 3.74TTEE43 pKa = 4.0EE44 pKa = 3.97ADD46 pKa = 3.45AAPAEE51 pKa = 4.21LDD53 pKa = 3.27YY54 pKa = 11.77DD55 pKa = 4.03GDD57 pKa = 3.83IKK59 pKa = 11.41VWVADD64 pKa = 3.61NAVSFTQEE72 pKa = 3.6QIAKK76 pKa = 9.18FQEE79 pKa = 4.48ANPGVANATFTVEE92 pKa = 4.37AVGEE96 pKa = 4.0GDD98 pKa = 3.24AASNVITDD106 pKa = 3.45VKK108 pKa = 11.02AGGDD112 pKa = 3.62VYY114 pKa = 11.09TFVQDD119 pKa = 3.23QIARR123 pKa = 11.84FVAAGALEE131 pKa = 4.24EE132 pKa = 4.17VAPEE136 pKa = 3.64NVAAVEE142 pKa = 3.97ADD144 pKa = 3.39NDD146 pKa = 3.7EE147 pKa = 4.6GAVGAATVGSTLYY160 pKa = 10.47AYY162 pKa = 9.37PLTSDD167 pKa = 3.07NGYY170 pKa = 10.88FMYY173 pKa = 10.53YY174 pKa = 10.07DD175 pKa = 3.62KK176 pKa = 11.45SVVTDD181 pKa = 3.76PTDD184 pKa = 3.64LDD186 pKa = 5.22AIIADD191 pKa = 4.07CEE193 pKa = 4.16KK194 pKa = 10.61AGKK197 pKa = 9.08NIYY200 pKa = 9.33MEE202 pKa = 5.05INSGWYY208 pKa = 6.59QTAFFFGTGCDD219 pKa = 3.53LSYY222 pKa = 10.38EE223 pKa = 4.13TDD225 pKa = 3.41DD226 pKa = 4.57AGNFTSANVNYY237 pKa = 10.49ASDD240 pKa = 3.5QGLVALKK247 pKa = 10.63EE248 pKa = 4.21MIRR251 pKa = 11.84LSSSSAFVNGSAAGEE266 pKa = 4.08ATTIGAIVDD275 pKa = 4.22GTWDD279 pKa = 3.27ATIVKK284 pKa = 9.7EE285 pKa = 4.7VFGDD289 pKa = 3.74NYY291 pKa = 10.98GCTPLPSFQGSDD303 pKa = 2.67GKK305 pKa = 9.25TYY307 pKa = 10.88QMSGFSGYY315 pKa = 10.68KK316 pKa = 9.92LLGVKK321 pKa = 9.45PQEE324 pKa = 4.73DD325 pKa = 4.09EE326 pKa = 4.24NKK328 pKa = 10.59LAVCDD333 pKa = 4.12ALAAYY338 pKa = 9.69LSSGEE343 pKa = 4.17VQLARR348 pKa = 11.84YY349 pKa = 7.46EE350 pKa = 4.34AIGWGPSNLEE360 pKa = 3.81AQQSDD365 pKa = 3.96EE366 pKa = 4.27VQADD370 pKa = 3.76EE371 pKa = 5.53ALSALNEE378 pKa = 3.78QLLRR382 pKa = 11.84NKK384 pKa = 10.01PQGQYY389 pKa = 10.31PGDD392 pKa = 3.74YY393 pKa = 7.35WTLATGLGDD402 pKa = 5.46DD403 pKa = 5.28IITGTLNEE411 pKa = 4.37KK412 pKa = 10.74SSDD415 pKa = 3.9EE416 pKa = 4.28DD417 pKa = 4.55LMAALQTFQDD427 pKa = 3.63TCISYY432 pKa = 11.24AKK434 pKa = 10.53
MM1 pKa = 7.81KK2 pKa = 10.32KK3 pKa = 10.3RR4 pKa = 11.84IVSVLMAGALASAMLAGCGNSSAPAASAAAEE35 pKa = 4.18DD36 pKa = 4.01TAEE39 pKa = 4.36DD40 pKa = 3.74TTEE43 pKa = 4.0EE44 pKa = 3.97ADD46 pKa = 3.45AAPAEE51 pKa = 4.21LDD53 pKa = 3.27YY54 pKa = 11.77DD55 pKa = 4.03GDD57 pKa = 3.83IKK59 pKa = 11.41VWVADD64 pKa = 3.61NAVSFTQEE72 pKa = 3.6QIAKK76 pKa = 9.18FQEE79 pKa = 4.48ANPGVANATFTVEE92 pKa = 4.37AVGEE96 pKa = 4.0GDD98 pKa = 3.24AASNVITDD106 pKa = 3.45VKK108 pKa = 11.02AGGDD112 pKa = 3.62VYY114 pKa = 11.09TFVQDD119 pKa = 3.23QIARR123 pKa = 11.84FVAAGALEE131 pKa = 4.24EE132 pKa = 4.17VAPEE136 pKa = 3.64NVAAVEE142 pKa = 3.97ADD144 pKa = 3.39NDD146 pKa = 3.7EE147 pKa = 4.6GAVGAATVGSTLYY160 pKa = 10.47AYY162 pKa = 9.37PLTSDD167 pKa = 3.07NGYY170 pKa = 10.88FMYY173 pKa = 10.53YY174 pKa = 10.07DD175 pKa = 3.62KK176 pKa = 11.45SVVTDD181 pKa = 3.76PTDD184 pKa = 3.64LDD186 pKa = 5.22AIIADD191 pKa = 4.07CEE193 pKa = 4.16KK194 pKa = 10.61AGKK197 pKa = 9.08NIYY200 pKa = 9.33MEE202 pKa = 5.05INSGWYY208 pKa = 6.59QTAFFFGTGCDD219 pKa = 3.53LSYY222 pKa = 10.38EE223 pKa = 4.13TDD225 pKa = 3.41DD226 pKa = 4.57AGNFTSANVNYY237 pKa = 10.49ASDD240 pKa = 3.5QGLVALKK247 pKa = 10.63EE248 pKa = 4.21MIRR251 pKa = 11.84LSSSSAFVNGSAAGEE266 pKa = 4.08ATTIGAIVDD275 pKa = 4.22GTWDD279 pKa = 3.27ATIVKK284 pKa = 9.7EE285 pKa = 4.7VFGDD289 pKa = 3.74NYY291 pKa = 10.98GCTPLPSFQGSDD303 pKa = 2.67GKK305 pKa = 9.25TYY307 pKa = 10.88QMSGFSGYY315 pKa = 10.68KK316 pKa = 9.92LLGVKK321 pKa = 9.45PQEE324 pKa = 4.73DD325 pKa = 4.09EE326 pKa = 4.24NKK328 pKa = 10.59LAVCDD333 pKa = 4.12ALAAYY338 pKa = 9.69LSSGEE343 pKa = 4.17VQLARR348 pKa = 11.84YY349 pKa = 7.46EE350 pKa = 4.34AIGWGPSNLEE360 pKa = 3.81AQQSDD365 pKa = 3.96EE366 pKa = 4.27VQADD370 pKa = 3.76EE371 pKa = 5.53ALSALNEE378 pKa = 3.78QLLRR382 pKa = 11.84NKK384 pKa = 10.01PQGQYY389 pKa = 10.31PGDD392 pKa = 3.74YY393 pKa = 7.35WTLATGLGDD402 pKa = 5.46DD403 pKa = 5.28IITGTLNEE411 pKa = 4.37KK412 pKa = 10.74SSDD415 pKa = 3.9EE416 pKa = 4.28DD417 pKa = 4.55LMAALQTFQDD427 pKa = 3.63TCISYY432 pKa = 11.24AKK434 pKa = 10.53
Molecular weight: 45.63 kDa
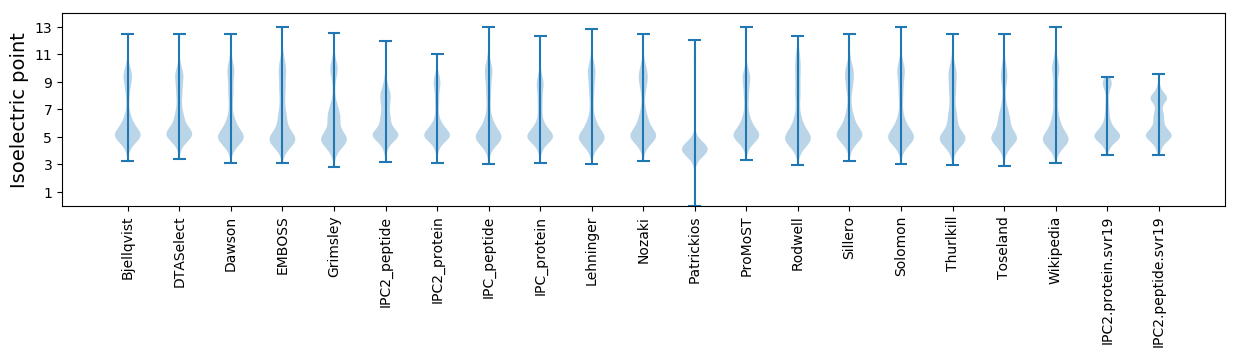
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5CA28|R5CA28_9FIRM Uncharacterized protein OS=Firmicutes bacterium CAG:791 OX=1262993 GN=BN785_00896 PE=4 SV=1
MM1 pKa = 7.72AKK3 pKa = 8.07MTYY6 pKa = 8.55QPKK9 pKa = 9.46KK10 pKa = 7.57RR11 pKa = 11.84QRR13 pKa = 11.84AKK15 pKa = 9.44VHH17 pKa = 5.53GFRR20 pKa = 11.84ARR22 pKa = 11.84MASAGGRR29 pKa = 11.84KK30 pKa = 8.85VLAARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.11GRR40 pKa = 11.84KK41 pKa = 8.21QLTVV45 pKa = 2.97
MM1 pKa = 7.72AKK3 pKa = 8.07MTYY6 pKa = 8.55QPKK9 pKa = 9.46KK10 pKa = 7.57RR11 pKa = 11.84QRR13 pKa = 11.84AKK15 pKa = 9.44VHH17 pKa = 5.53GFRR20 pKa = 11.84ARR22 pKa = 11.84MASAGGRR29 pKa = 11.84KK30 pKa = 8.85VLAARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.11GRR40 pKa = 11.84KK41 pKa = 8.21QLTVV45 pKa = 2.97
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
699191 |
29 |
1537 |
321.5 |
35.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.438 ± 0.061 | 1.55 ± 0.022 |
5.759 ± 0.05 | 7.417 ± 0.065 |
4.076 ± 0.036 | 7.489 ± 0.046 |
2.007 ± 0.025 | 6.468 ± 0.051 |
5.518 ± 0.041 | 9.455 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.021 ± 0.026 | 3.786 ± 0.03 |
3.692 ± 0.03 | 3.269 ± 0.027 |
5.497 ± 0.051 | 5.869 ± 0.039 |
5.178 ± 0.038 | 6.733 ± 0.044 |
0.955 ± 0.017 | 3.821 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
