
Wuhan house centipede virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.35
Get precalculated fractions of proteins
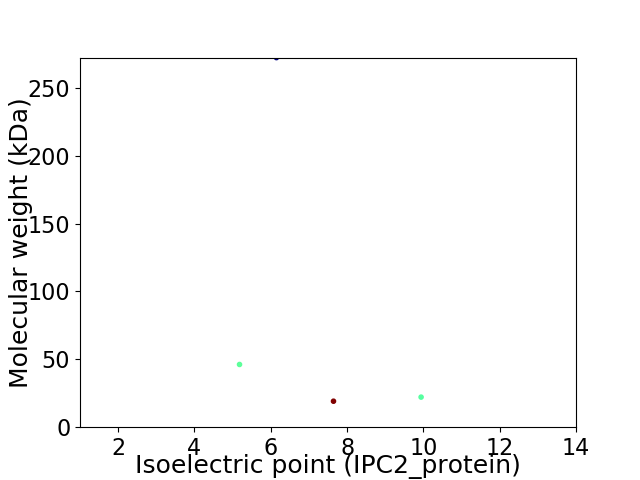
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
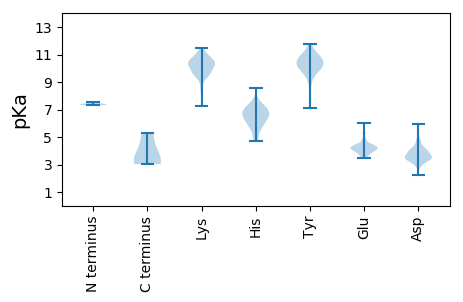
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KKG1|A0A1L3KKG1_9VIRU Non-structural protein 3 OS=Wuhan house centipede virus 1 OX=1923705 PE=4 SV=1
MM1 pKa = 7.32YY2 pKa = 10.36IKK4 pKa = 10.56FCVLLYY10 pKa = 9.95FVSFVNSAQLYY21 pKa = 9.49SRR23 pKa = 11.84WLEE26 pKa = 3.61NGYY29 pKa = 10.47FGTNITLPLLSLKK42 pKa = 9.31YY43 pKa = 9.08TEE45 pKa = 4.76TPVGQYY51 pKa = 10.15SRR53 pKa = 11.84SPFVFEE59 pKa = 4.44KK60 pKa = 10.82SCISVVKK67 pKa = 9.99TNTLLEE73 pKa = 4.24VDD75 pKa = 4.16CQLPQHH81 pKa = 6.53CGVLHH86 pKa = 6.03HH87 pKa = 7.16RR88 pKa = 11.84KK89 pKa = 8.24MPQGWFGQEE98 pKa = 3.85VILCYY103 pKa = 10.45SFHH106 pKa = 5.66QTTLGGALKK115 pKa = 10.55VSTMDD120 pKa = 3.79FSPSSFTYY128 pKa = 9.85EE129 pKa = 4.25SPNGPLTVEE138 pKa = 4.47DD139 pKa = 3.72WTSVDD144 pKa = 4.29FFSHH148 pKa = 6.9VYY150 pKa = 10.4LLNSNDD156 pKa = 3.81RR157 pKa = 11.84YY158 pKa = 11.2SLIPRR163 pKa = 11.84ICLDD167 pKa = 4.12HH168 pKa = 5.96YY169 pKa = 10.01TSSNDD174 pKa = 3.58DD175 pKa = 3.41QPQSVQFEE183 pKa = 4.29FTEE186 pKa = 4.34SQVCVHH192 pKa = 6.06QSLHH196 pKa = 6.84PEE198 pKa = 4.14TDD200 pKa = 4.73CEE202 pKa = 4.4PEE204 pKa = 3.87WYY206 pKa = 10.1EE207 pKa = 4.61SMEE210 pKa = 4.07KK211 pKa = 10.7LIVSYY216 pKa = 9.84DD217 pKa = 3.41YY218 pKa = 11.0PVAYY222 pKa = 10.09RR223 pKa = 11.84GNPYY227 pKa = 10.68EE228 pKa = 4.25LVTHH232 pKa = 6.48IRR234 pKa = 11.84YY235 pKa = 9.67GNQDD239 pKa = 3.72PNVINYY245 pKa = 8.73VSSNKK250 pKa = 9.59LNHH253 pKa = 5.72VFSVIDD259 pKa = 3.71GKK261 pKa = 9.46TYY263 pKa = 10.7SSAVGLSHH271 pKa = 7.08MYY273 pKa = 10.51LHH275 pKa = 7.42GISSPEE281 pKa = 3.81NYY283 pKa = 9.13IVLVDD288 pKa = 4.14PSPLKK293 pKa = 10.86VNQSFCYY300 pKa = 9.79PIQSVYY306 pKa = 10.88KK307 pKa = 9.13NPLSAVINSIIDD319 pKa = 3.39ALEE322 pKa = 4.4PIILKK327 pKa = 10.18LLSILEE333 pKa = 4.07QYY335 pKa = 10.57LEE337 pKa = 4.19KK338 pKa = 10.4FLKK341 pKa = 10.45IFIKK345 pKa = 10.76ALLIFLKK352 pKa = 10.61LFYY355 pKa = 10.77EE356 pKa = 5.33LIDD359 pKa = 3.68NLLTQEE365 pKa = 4.9IITALLCSAAVYY377 pKa = 9.92LYY379 pKa = 10.64FKK381 pKa = 10.44DD382 pKa = 3.95YY383 pKa = 11.14VITFISFCLFCFLLYY398 pKa = 10.85YY399 pKa = 10.22IFF401 pKa = 5.33
MM1 pKa = 7.32YY2 pKa = 10.36IKK4 pKa = 10.56FCVLLYY10 pKa = 9.95FVSFVNSAQLYY21 pKa = 9.49SRR23 pKa = 11.84WLEE26 pKa = 3.61NGYY29 pKa = 10.47FGTNITLPLLSLKK42 pKa = 9.31YY43 pKa = 9.08TEE45 pKa = 4.76TPVGQYY51 pKa = 10.15SRR53 pKa = 11.84SPFVFEE59 pKa = 4.44KK60 pKa = 10.82SCISVVKK67 pKa = 9.99TNTLLEE73 pKa = 4.24VDD75 pKa = 4.16CQLPQHH81 pKa = 6.53CGVLHH86 pKa = 6.03HH87 pKa = 7.16RR88 pKa = 11.84KK89 pKa = 8.24MPQGWFGQEE98 pKa = 3.85VILCYY103 pKa = 10.45SFHH106 pKa = 5.66QTTLGGALKK115 pKa = 10.55VSTMDD120 pKa = 3.79FSPSSFTYY128 pKa = 9.85EE129 pKa = 4.25SPNGPLTVEE138 pKa = 4.47DD139 pKa = 3.72WTSVDD144 pKa = 4.29FFSHH148 pKa = 6.9VYY150 pKa = 10.4LLNSNDD156 pKa = 3.81RR157 pKa = 11.84YY158 pKa = 11.2SLIPRR163 pKa = 11.84ICLDD167 pKa = 4.12HH168 pKa = 5.96YY169 pKa = 10.01TSSNDD174 pKa = 3.58DD175 pKa = 3.41QPQSVQFEE183 pKa = 4.29FTEE186 pKa = 4.34SQVCVHH192 pKa = 6.06QSLHH196 pKa = 6.84PEE198 pKa = 4.14TDD200 pKa = 4.73CEE202 pKa = 4.4PEE204 pKa = 3.87WYY206 pKa = 10.1EE207 pKa = 4.61SMEE210 pKa = 4.07KK211 pKa = 10.7LIVSYY216 pKa = 9.84DD217 pKa = 3.41YY218 pKa = 11.0PVAYY222 pKa = 10.09RR223 pKa = 11.84GNPYY227 pKa = 10.68EE228 pKa = 4.25LVTHH232 pKa = 6.48IRR234 pKa = 11.84YY235 pKa = 9.67GNQDD239 pKa = 3.72PNVINYY245 pKa = 8.73VSSNKK250 pKa = 9.59LNHH253 pKa = 5.72VFSVIDD259 pKa = 3.71GKK261 pKa = 9.46TYY263 pKa = 10.7SSAVGLSHH271 pKa = 7.08MYY273 pKa = 10.51LHH275 pKa = 7.42GISSPEE281 pKa = 3.81NYY283 pKa = 9.13IVLVDD288 pKa = 4.14PSPLKK293 pKa = 10.86VNQSFCYY300 pKa = 9.79PIQSVYY306 pKa = 10.88KK307 pKa = 9.13NPLSAVINSIIDD319 pKa = 3.39ALEE322 pKa = 4.4PIILKK327 pKa = 10.18LLSILEE333 pKa = 4.07QYY335 pKa = 10.57LEE337 pKa = 4.19KK338 pKa = 10.4FLKK341 pKa = 10.45IFIKK345 pKa = 10.76ALLIFLKK352 pKa = 10.61LFYY355 pKa = 10.77EE356 pKa = 5.33LIDD359 pKa = 3.68NLLTQEE365 pKa = 4.9IITALLCSAAVYY377 pKa = 9.92LYY379 pKa = 10.64FKK381 pKa = 10.44DD382 pKa = 3.95YY383 pKa = 11.14VITFISFCLFCFLLYY398 pKa = 10.85YY399 pKa = 10.22IFF401 pKa = 5.33
Molecular weight: 46.22 kDa
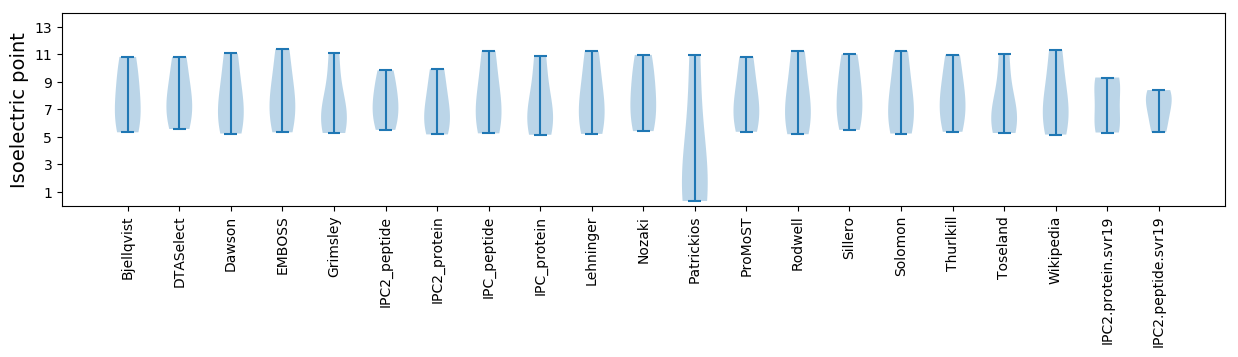
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KKD7|A0A1L3KKD7_9VIRU DiSB-ORF2_chro domain-containing protein OS=Wuhan house centipede virus 1 OX=1923705 PE=4 SV=1
MM1 pKa = 7.39SSAVRR6 pKa = 11.84NRR8 pKa = 11.84SRR10 pKa = 11.84IPVVSQRR17 pKa = 11.84TVQRR21 pKa = 11.84PRR23 pKa = 11.84VVTRR27 pKa = 11.84TRR29 pKa = 11.84VVVPRR34 pKa = 11.84QKK36 pKa = 9.46TRR38 pKa = 11.84NTPRR42 pKa = 11.84SNYY45 pKa = 9.49FDD47 pKa = 4.14LNKK50 pKa = 9.82VFTDD54 pKa = 3.59ISSTFTRR61 pKa = 11.84ALSNPLVLCTISIALGVILTHH82 pKa = 7.55DD83 pKa = 3.94FEE85 pKa = 5.25KK86 pKa = 11.06KK87 pKa = 9.94NGYY90 pKa = 9.84IYY92 pKa = 9.59DD93 pKa = 3.91TFKK96 pKa = 11.05EE97 pKa = 4.19RR98 pKa = 11.84NDD100 pKa = 4.84NISQWIVNNGKK111 pKa = 9.59KK112 pKa = 9.93FAGLLIFAPAVVDD125 pKa = 4.03APRR128 pKa = 11.84NLRR131 pKa = 11.84AVVALVSFLWVMIIPEE147 pKa = 4.31SKK149 pKa = 9.13AAEE152 pKa = 4.1YY153 pKa = 10.19FLQSLALHH161 pKa = 6.44TYY163 pKa = 9.85FRR165 pKa = 11.84LSNNNSKK172 pKa = 10.8LMVLFMVVIAWFLGFLTVGKK192 pKa = 10.22NSSS195 pKa = 3.04
MM1 pKa = 7.39SSAVRR6 pKa = 11.84NRR8 pKa = 11.84SRR10 pKa = 11.84IPVVSQRR17 pKa = 11.84TVQRR21 pKa = 11.84PRR23 pKa = 11.84VVTRR27 pKa = 11.84TRR29 pKa = 11.84VVVPRR34 pKa = 11.84QKK36 pKa = 9.46TRR38 pKa = 11.84NTPRR42 pKa = 11.84SNYY45 pKa = 9.49FDD47 pKa = 4.14LNKK50 pKa = 9.82VFTDD54 pKa = 3.59ISSTFTRR61 pKa = 11.84ALSNPLVLCTISIALGVILTHH82 pKa = 7.55DD83 pKa = 3.94FEE85 pKa = 5.25KK86 pKa = 11.06KK87 pKa = 9.94NGYY90 pKa = 9.84IYY92 pKa = 9.59DD93 pKa = 3.91TFKK96 pKa = 11.05EE97 pKa = 4.19RR98 pKa = 11.84NDD100 pKa = 4.84NISQWIVNNGKK111 pKa = 9.59KK112 pKa = 9.93FAGLLIFAPAVVDD125 pKa = 4.03APRR128 pKa = 11.84NLRR131 pKa = 11.84AVVALVSFLWVMIIPEE147 pKa = 4.31SKK149 pKa = 9.13AAEE152 pKa = 4.1YY153 pKa = 10.19FLQSLALHH161 pKa = 6.44TYY163 pKa = 9.85FRR165 pKa = 11.84LSNNNSKK172 pKa = 10.8LMVLFMVVIAWFLGFLTVGKK192 pKa = 10.22NSSS195 pKa = 3.04
Molecular weight: 22.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3135 |
166 |
2373 |
783.8 |
89.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.402 ± 0.54 | 2.711 ± 0.363 |
5.04 ± 0.682 | 5.71 ± 1.092 |
5.423 ± 0.605 | 3.7 ± 0.211 |
2.679 ± 0.256 | 6.252 ± 0.234 |
5.391 ± 0.552 | 9.601 ± 0.884 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.978 ± 0.285 | 5.837 ± 0.582 |
4.402 ± 0.265 | 3.126 ± 0.307 |
5.136 ± 0.898 | 8.995 ± 0.459 |
5.678 ± 0.203 | 7.719 ± 0.729 |
0.574 ± 0.396 | 5.646 ± 0.586 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
