
Smithella sp. D17
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Syntrophobacterales; Syntrophaceae; Smithella; unclassified Smithella
Average proteome isoelectric point is 7.21
Get precalculated fractions of proteins
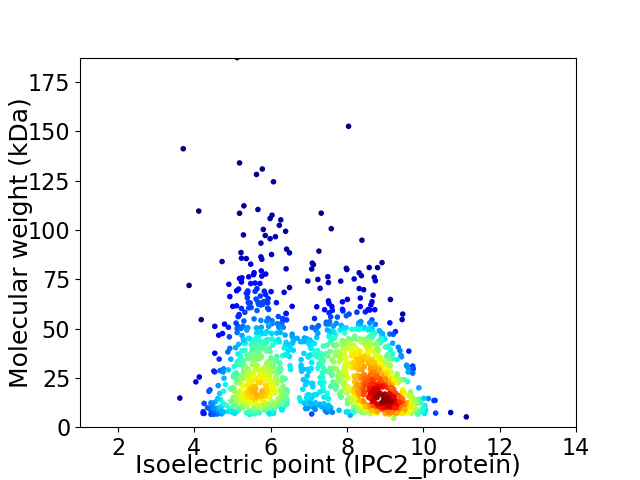
Virtual 2D-PAGE plot for 1343 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A094K1Q2|A0A094K1Q2_9DELT Antitoxin OS=Smithella sp. D17 OX=1538639 GN=KD27_07510 PE=4 SV=1
MM1 pKa = 7.97AEE3 pKa = 3.67IKK5 pKa = 10.76KK6 pKa = 10.06PVTSEE11 pKa = 3.59MGHH14 pKa = 4.9QVTGKK19 pKa = 10.24VAILYY24 pKa = 9.89GNVKK28 pKa = 10.3AISPDD33 pKa = 2.91GTVRR37 pKa = 11.84MLKK40 pKa = 10.09INSPVFADD48 pKa = 4.05DD49 pKa = 5.01RR50 pKa = 11.84IITGGDD56 pKa = 3.2GSVSIVFNTSPSTQLDD72 pKa = 3.7LGRR75 pKa = 11.84VSDD78 pKa = 3.81MVIDD82 pKa = 3.74EE83 pKa = 4.4DD84 pKa = 4.74VYY86 pKa = 10.91GVVSKK91 pKa = 10.83GVAAEE96 pKa = 4.29ASAEE100 pKa = 3.9QEE102 pKa = 4.48AIQKK106 pKa = 10.5ALLAGDD112 pKa = 3.75QPLEE116 pKa = 4.25LDD118 pKa = 3.29ATAAGAEE125 pKa = 4.13ANAGGGHH132 pKa = 6.52PVFVVTPDD140 pKa = 2.98WSEE143 pKa = 3.85VTPEE147 pKa = 3.95SGAEE151 pKa = 3.85TRR153 pKa = 11.84GITWGTGVSTNYY165 pKa = 10.41DD166 pKa = 3.69PNLDD170 pKa = 3.64QPITVSISDD179 pKa = 4.48GLPDD183 pKa = 4.4PQSEE187 pKa = 4.5GQGGNVITFTVSLSEE202 pKa = 4.13ATNHH206 pKa = 6.14DD207 pKa = 3.42TTVNYY212 pKa = 7.33TTADD216 pKa = 3.45GTAVAGTDD224 pKa = 3.67YY225 pKa = 10.66IATSGTITFLAGEE238 pKa = 4.36TTATVTVPVLNDD250 pKa = 3.46TVFEE254 pKa = 4.16NPEE257 pKa = 3.92AFTVQLGTPSTGVTIVDD274 pKa = 3.94GSGIGNIADD283 pKa = 3.72NDD285 pKa = 3.94AVPAFSINDD294 pKa = 3.36VTVTEE299 pKa = 4.87GTDD302 pKa = 3.24STITFTVTKK311 pKa = 10.1TGLTDD316 pKa = 3.21QASSVDD322 pKa = 3.79YY323 pKa = 11.11SVAPNTAVTPGDD335 pKa = 3.68YY336 pKa = 8.95TASDD340 pKa = 4.32LLAGTLTFAAGEE352 pKa = 4.29TTKK355 pKa = 10.64TITLNIADD363 pKa = 4.16DD364 pKa = 3.86SVYY367 pKa = 11.07EE368 pKa = 3.97LTEE371 pKa = 4.01TFNVNLSNAAAATISDD387 pKa = 3.88TQGVGTILDD396 pKa = 4.21NDD398 pKa = 4.15DD399 pKa = 3.81TDD401 pKa = 3.69PTAGIIYY408 pKa = 8.22ATVDD412 pKa = 3.52DD413 pKa = 5.01EE414 pKa = 4.55GLSGGISGGTGDD426 pKa = 4.76IVVTPDD432 pKa = 3.64PDD434 pKa = 4.06SNEE437 pKa = 3.79ATFTGILTHH446 pKa = 7.22DD447 pKa = 4.43FVLNGPGSIGFASMNGLTGIVGQEE471 pKa = 3.83TVTYY475 pKa = 9.07SWTGSTLTATGPRR488 pKa = 11.84GALFTVEE495 pKa = 4.22VTDD498 pKa = 4.33PATGAYY504 pKa = 9.34KK505 pKa = 9.48VTLLDD510 pKa = 3.73NVLHH514 pKa = 6.07GTLDD518 pKa = 3.68GQVGDD523 pKa = 3.78NTEE526 pKa = 4.21NNATAALNYY535 pKa = 8.76TVTDD539 pKa = 3.84ADD541 pKa = 4.01GDD543 pKa = 4.31TATGTLNITFNDD555 pKa = 4.12DD556 pKa = 3.1MPTANDD562 pKa = 3.23QSVTVVVHH570 pKa = 7.03DD571 pKa = 4.23SATTAHH577 pKa = 6.97ISALAAGWVAPFDD590 pKa = 3.98INVDD594 pKa = 3.76TKK596 pKa = 10.81TSTDD600 pKa = 3.03ADD602 pKa = 4.02TYY604 pKa = 10.42FEE606 pKa = 4.73KK607 pKa = 10.61IAWGDD612 pKa = 3.49NSNSSSYY619 pKa = 11.17VYY621 pKa = 11.0NDD623 pKa = 3.01NNALATVNVGEE634 pKa = 4.45TFTLGTFTHH643 pKa = 6.54NNFPIPSGSSITDD656 pKa = 3.3AYY658 pKa = 11.27LKK660 pKa = 9.57VTFDD664 pKa = 3.79VVIDD668 pKa = 4.1GQTVHH673 pKa = 6.11VDD675 pKa = 3.2KK676 pKa = 10.63TVHH679 pKa = 5.58FQHH682 pKa = 6.9NEE684 pKa = 3.95TPNDD688 pKa = 3.68GANPDD693 pKa = 4.5DD694 pKa = 4.26IVTIVNGTNVVDD706 pKa = 4.01VKK708 pKa = 11.38VGDD711 pKa = 3.64FTYY714 pKa = 10.51TLTIGFQDD722 pKa = 3.2ASGNTVTQVYY732 pKa = 9.24TDD734 pKa = 3.67EE735 pKa = 4.55SAANTFNLNATLTADD750 pKa = 3.77ADD752 pKa = 4.04HH753 pKa = 7.04YY754 pKa = 10.54ILPVVGTVDD763 pKa = 3.22ASFGADD769 pKa = 3.25GPGALHH775 pKa = 6.53IVSIAHH781 pKa = 6.99DD782 pKa = 4.21ANGDD786 pKa = 4.02GIDD789 pKa = 3.6EE790 pKa = 4.61VYY792 pKa = 9.28NTSSGGYY799 pKa = 9.73NSTTTTLTIATHH811 pKa = 6.11EE812 pKa = 4.66GGTLAVNFSTGAYY825 pKa = 7.92TYY827 pKa = 7.37TAPTGTPLVANEE839 pKa = 3.93VFTYY843 pKa = 10.37TIMDD847 pKa = 4.24ADD849 pKa = 4.5GDD851 pKa = 4.17TATANLTVNLPVHH864 pKa = 6.89DD865 pKa = 4.16MLVVGTNADD874 pKa = 3.76DD875 pKa = 3.83VTGQTIDD882 pKa = 3.23HH883 pKa = 6.98HH884 pKa = 7.07IDD886 pKa = 3.03HH887 pKa = 6.94YY888 pKa = 11.61SPLDD892 pKa = 3.57GPITGGGGNDD902 pKa = 3.5VLVGDD907 pKa = 4.58IGGANNIVQPGQNYY921 pKa = 9.98NISLIVDD928 pKa = 3.83SSGSMDD934 pKa = 4.13KK935 pKa = 11.15DD936 pKa = 3.34SGTAGYY942 pKa = 8.38TRR944 pKa = 11.84MEE946 pKa = 4.46LAQAALKK953 pKa = 10.63NLADD957 pKa = 3.67QLVGHH962 pKa = 7.52DD963 pKa = 3.92GTINLQIIDD972 pKa = 3.84FDD974 pKa = 5.63SNVVANTSWSNITSANLSAIYY995 pKa = 8.71TAIDD999 pKa = 3.25NMVAEE1004 pKa = 4.9GATNYY1009 pKa = 8.82EE1010 pKa = 4.03AGFNASNAWLSSQTNGYY1027 pKa = 10.04EE1028 pKa = 3.98NITFFLTDD1036 pKa = 4.1GNPTYY1041 pKa = 10.78YY1042 pKa = 10.0INSYY1046 pKa = 10.85GNVAGPGKK1054 pKa = 9.3VTDD1057 pKa = 4.32YY1058 pKa = 10.4YY1059 pKa = 11.52TMLNSVEE1066 pKa = 4.61AFALLSPITHH1076 pKa = 6.09VEE1078 pKa = 4.08AIGIGSEE1085 pKa = 4.12VNEE1088 pKa = 4.29NYY1090 pKa = 10.79LKK1092 pKa = 10.9FFDD1095 pKa = 3.99NTDD1098 pKa = 3.03VTTEE1102 pKa = 3.95TGSVSFWNGTVTGPVGQVEE1121 pKa = 4.41IVHH1124 pKa = 5.6TAEE1127 pKa = 4.16EE1128 pKa = 4.04LSAALQHH1135 pKa = 6.8GSLEE1139 pKa = 4.2YY1140 pKa = 10.15TLLGAGDD1147 pKa = 4.08DD1148 pKa = 4.35HH1149 pKa = 6.83LASGDD1154 pKa = 3.54GDD1156 pKa = 3.94DD1157 pKa = 5.44VIFGDD1162 pKa = 5.18SVHH1165 pKa = 6.64TDD1167 pKa = 3.05ALADD1171 pKa = 4.11ANPSLTTADD1180 pKa = 3.48GAGWEE1185 pKa = 4.33VFEE1188 pKa = 4.34KK1189 pKa = 10.97LGWTEE1194 pKa = 3.87QQITDD1199 pKa = 4.06YY1200 pKa = 10.94IKK1202 pKa = 10.98ANHH1205 pKa = 5.87STLATEE1211 pKa = 4.55SGRR1214 pKa = 11.84TGGNDD1219 pKa = 3.69TIDD1222 pKa = 3.56GGAGNDD1228 pKa = 3.39IIYY1231 pKa = 9.65GQEE1234 pKa = 3.95GNDD1237 pKa = 3.65VINGGEE1243 pKa = 4.26GNDD1246 pKa = 4.03LLSGGTGNNILNGGLGADD1264 pKa = 3.45TFVISKK1270 pKa = 9.99GGHH1273 pKa = 5.73DD1274 pKa = 4.59TIQDD1278 pKa = 3.77YY1279 pKa = 11.58NKK1281 pKa = 10.6GQLDD1285 pKa = 3.73KK1286 pKa = 11.42VDD1288 pKa = 3.19ISSIVKK1294 pKa = 8.57VTDD1297 pKa = 3.44DD1298 pKa = 4.26AVAKK1302 pKa = 9.43TYY1304 pKa = 10.96LGVEE1308 pKa = 3.89QNANGKK1314 pKa = 9.48AVLDD1318 pKa = 4.04IYY1320 pKa = 11.61DD1321 pKa = 3.91NGSDD1325 pKa = 3.29HH1326 pKa = 7.29SAGHH1330 pKa = 6.72LVATVTFDD1338 pKa = 3.66NIHH1341 pKa = 6.05YY1342 pKa = 9.91SDD1344 pKa = 5.1LGSIHH1349 pKa = 6.51QLDD1352 pKa = 4.21SLLGAVDD1359 pKa = 3.91INHH1362 pKa = 6.23SS1363 pKa = 3.57
MM1 pKa = 7.97AEE3 pKa = 3.67IKK5 pKa = 10.76KK6 pKa = 10.06PVTSEE11 pKa = 3.59MGHH14 pKa = 4.9QVTGKK19 pKa = 10.24VAILYY24 pKa = 9.89GNVKK28 pKa = 10.3AISPDD33 pKa = 2.91GTVRR37 pKa = 11.84MLKK40 pKa = 10.09INSPVFADD48 pKa = 4.05DD49 pKa = 5.01RR50 pKa = 11.84IITGGDD56 pKa = 3.2GSVSIVFNTSPSTQLDD72 pKa = 3.7LGRR75 pKa = 11.84VSDD78 pKa = 3.81MVIDD82 pKa = 3.74EE83 pKa = 4.4DD84 pKa = 4.74VYY86 pKa = 10.91GVVSKK91 pKa = 10.83GVAAEE96 pKa = 4.29ASAEE100 pKa = 3.9QEE102 pKa = 4.48AIQKK106 pKa = 10.5ALLAGDD112 pKa = 3.75QPLEE116 pKa = 4.25LDD118 pKa = 3.29ATAAGAEE125 pKa = 4.13ANAGGGHH132 pKa = 6.52PVFVVTPDD140 pKa = 2.98WSEE143 pKa = 3.85VTPEE147 pKa = 3.95SGAEE151 pKa = 3.85TRR153 pKa = 11.84GITWGTGVSTNYY165 pKa = 10.41DD166 pKa = 3.69PNLDD170 pKa = 3.64QPITVSISDD179 pKa = 4.48GLPDD183 pKa = 4.4PQSEE187 pKa = 4.5GQGGNVITFTVSLSEE202 pKa = 4.13ATNHH206 pKa = 6.14DD207 pKa = 3.42TTVNYY212 pKa = 7.33TTADD216 pKa = 3.45GTAVAGTDD224 pKa = 3.67YY225 pKa = 10.66IATSGTITFLAGEE238 pKa = 4.36TTATVTVPVLNDD250 pKa = 3.46TVFEE254 pKa = 4.16NPEE257 pKa = 3.92AFTVQLGTPSTGVTIVDD274 pKa = 3.94GSGIGNIADD283 pKa = 3.72NDD285 pKa = 3.94AVPAFSINDD294 pKa = 3.36VTVTEE299 pKa = 4.87GTDD302 pKa = 3.24STITFTVTKK311 pKa = 10.1TGLTDD316 pKa = 3.21QASSVDD322 pKa = 3.79YY323 pKa = 11.11SVAPNTAVTPGDD335 pKa = 3.68YY336 pKa = 8.95TASDD340 pKa = 4.32LLAGTLTFAAGEE352 pKa = 4.29TTKK355 pKa = 10.64TITLNIADD363 pKa = 4.16DD364 pKa = 3.86SVYY367 pKa = 11.07EE368 pKa = 3.97LTEE371 pKa = 4.01TFNVNLSNAAAATISDD387 pKa = 3.88TQGVGTILDD396 pKa = 4.21NDD398 pKa = 4.15DD399 pKa = 3.81TDD401 pKa = 3.69PTAGIIYY408 pKa = 8.22ATVDD412 pKa = 3.52DD413 pKa = 5.01EE414 pKa = 4.55GLSGGISGGTGDD426 pKa = 4.76IVVTPDD432 pKa = 3.64PDD434 pKa = 4.06SNEE437 pKa = 3.79ATFTGILTHH446 pKa = 7.22DD447 pKa = 4.43FVLNGPGSIGFASMNGLTGIVGQEE471 pKa = 3.83TVTYY475 pKa = 9.07SWTGSTLTATGPRR488 pKa = 11.84GALFTVEE495 pKa = 4.22VTDD498 pKa = 4.33PATGAYY504 pKa = 9.34KK505 pKa = 9.48VTLLDD510 pKa = 3.73NVLHH514 pKa = 6.07GTLDD518 pKa = 3.68GQVGDD523 pKa = 3.78NTEE526 pKa = 4.21NNATAALNYY535 pKa = 8.76TVTDD539 pKa = 3.84ADD541 pKa = 4.01GDD543 pKa = 4.31TATGTLNITFNDD555 pKa = 4.12DD556 pKa = 3.1MPTANDD562 pKa = 3.23QSVTVVVHH570 pKa = 7.03DD571 pKa = 4.23SATTAHH577 pKa = 6.97ISALAAGWVAPFDD590 pKa = 3.98INVDD594 pKa = 3.76TKK596 pKa = 10.81TSTDD600 pKa = 3.03ADD602 pKa = 4.02TYY604 pKa = 10.42FEE606 pKa = 4.73KK607 pKa = 10.61IAWGDD612 pKa = 3.49NSNSSSYY619 pKa = 11.17VYY621 pKa = 11.0NDD623 pKa = 3.01NNALATVNVGEE634 pKa = 4.45TFTLGTFTHH643 pKa = 6.54NNFPIPSGSSITDD656 pKa = 3.3AYY658 pKa = 11.27LKK660 pKa = 9.57VTFDD664 pKa = 3.79VVIDD668 pKa = 4.1GQTVHH673 pKa = 6.11VDD675 pKa = 3.2KK676 pKa = 10.63TVHH679 pKa = 5.58FQHH682 pKa = 6.9NEE684 pKa = 3.95TPNDD688 pKa = 3.68GANPDD693 pKa = 4.5DD694 pKa = 4.26IVTIVNGTNVVDD706 pKa = 4.01VKK708 pKa = 11.38VGDD711 pKa = 3.64FTYY714 pKa = 10.51TLTIGFQDD722 pKa = 3.2ASGNTVTQVYY732 pKa = 9.24TDD734 pKa = 3.67EE735 pKa = 4.55SAANTFNLNATLTADD750 pKa = 3.77ADD752 pKa = 4.04HH753 pKa = 7.04YY754 pKa = 10.54ILPVVGTVDD763 pKa = 3.22ASFGADD769 pKa = 3.25GPGALHH775 pKa = 6.53IVSIAHH781 pKa = 6.99DD782 pKa = 4.21ANGDD786 pKa = 4.02GIDD789 pKa = 3.6EE790 pKa = 4.61VYY792 pKa = 9.28NTSSGGYY799 pKa = 9.73NSTTTTLTIATHH811 pKa = 6.11EE812 pKa = 4.66GGTLAVNFSTGAYY825 pKa = 7.92TYY827 pKa = 7.37TAPTGTPLVANEE839 pKa = 3.93VFTYY843 pKa = 10.37TIMDD847 pKa = 4.24ADD849 pKa = 4.5GDD851 pKa = 4.17TATANLTVNLPVHH864 pKa = 6.89DD865 pKa = 4.16MLVVGTNADD874 pKa = 3.76DD875 pKa = 3.83VTGQTIDD882 pKa = 3.23HH883 pKa = 6.98HH884 pKa = 7.07IDD886 pKa = 3.03HH887 pKa = 6.94YY888 pKa = 11.61SPLDD892 pKa = 3.57GPITGGGGNDD902 pKa = 3.5VLVGDD907 pKa = 4.58IGGANNIVQPGQNYY921 pKa = 9.98NISLIVDD928 pKa = 3.83SSGSMDD934 pKa = 4.13KK935 pKa = 11.15DD936 pKa = 3.34SGTAGYY942 pKa = 8.38TRR944 pKa = 11.84MEE946 pKa = 4.46LAQAALKK953 pKa = 10.63NLADD957 pKa = 3.67QLVGHH962 pKa = 7.52DD963 pKa = 3.92GTINLQIIDD972 pKa = 3.84FDD974 pKa = 5.63SNVVANTSWSNITSANLSAIYY995 pKa = 8.71TAIDD999 pKa = 3.25NMVAEE1004 pKa = 4.9GATNYY1009 pKa = 8.82EE1010 pKa = 4.03AGFNASNAWLSSQTNGYY1027 pKa = 10.04EE1028 pKa = 3.98NITFFLTDD1036 pKa = 4.1GNPTYY1041 pKa = 10.78YY1042 pKa = 10.0INSYY1046 pKa = 10.85GNVAGPGKK1054 pKa = 9.3VTDD1057 pKa = 4.32YY1058 pKa = 10.4YY1059 pKa = 11.52TMLNSVEE1066 pKa = 4.61AFALLSPITHH1076 pKa = 6.09VEE1078 pKa = 4.08AIGIGSEE1085 pKa = 4.12VNEE1088 pKa = 4.29NYY1090 pKa = 10.79LKK1092 pKa = 10.9FFDD1095 pKa = 3.99NTDD1098 pKa = 3.03VTTEE1102 pKa = 3.95TGSVSFWNGTVTGPVGQVEE1121 pKa = 4.41IVHH1124 pKa = 5.6TAEE1127 pKa = 4.16EE1128 pKa = 4.04LSAALQHH1135 pKa = 6.8GSLEE1139 pKa = 4.2YY1140 pKa = 10.15TLLGAGDD1147 pKa = 4.08DD1148 pKa = 4.35HH1149 pKa = 6.83LASGDD1154 pKa = 3.54GDD1156 pKa = 3.94DD1157 pKa = 5.44VIFGDD1162 pKa = 5.18SVHH1165 pKa = 6.64TDD1167 pKa = 3.05ALADD1171 pKa = 4.11ANPSLTTADD1180 pKa = 3.48GAGWEE1185 pKa = 4.33VFEE1188 pKa = 4.34KK1189 pKa = 10.97LGWTEE1194 pKa = 3.87QQITDD1199 pKa = 4.06YY1200 pKa = 10.94IKK1202 pKa = 10.98ANHH1205 pKa = 5.87STLATEE1211 pKa = 4.55SGRR1214 pKa = 11.84TGGNDD1219 pKa = 3.69TIDD1222 pKa = 3.56GGAGNDD1228 pKa = 3.39IIYY1231 pKa = 9.65GQEE1234 pKa = 3.95GNDD1237 pKa = 3.65VINGGEE1243 pKa = 4.26GNDD1246 pKa = 4.03LLSGGTGNNILNGGLGADD1264 pKa = 3.45TFVISKK1270 pKa = 9.99GGHH1273 pKa = 5.73DD1274 pKa = 4.59TIQDD1278 pKa = 3.77YY1279 pKa = 11.58NKK1281 pKa = 10.6GQLDD1285 pKa = 3.73KK1286 pKa = 11.42VDD1288 pKa = 3.19ISSIVKK1294 pKa = 8.57VTDD1297 pKa = 3.44DD1298 pKa = 4.26AVAKK1302 pKa = 9.43TYY1304 pKa = 10.96LGVEE1308 pKa = 3.89QNANGKK1314 pKa = 9.48AVLDD1318 pKa = 4.04IYY1320 pKa = 11.61DD1321 pKa = 3.91NGSDD1325 pKa = 3.29HH1326 pKa = 7.29SAGHH1330 pKa = 6.72LVATVTFDD1338 pKa = 3.66NIHH1341 pKa = 6.05YY1342 pKa = 9.91SDD1344 pKa = 5.1LGSIHH1349 pKa = 6.51QLDD1352 pKa = 4.21SLLGAVDD1359 pKa = 3.91INHH1362 pKa = 6.23SS1363 pKa = 3.57
Molecular weight: 141.19 kDa
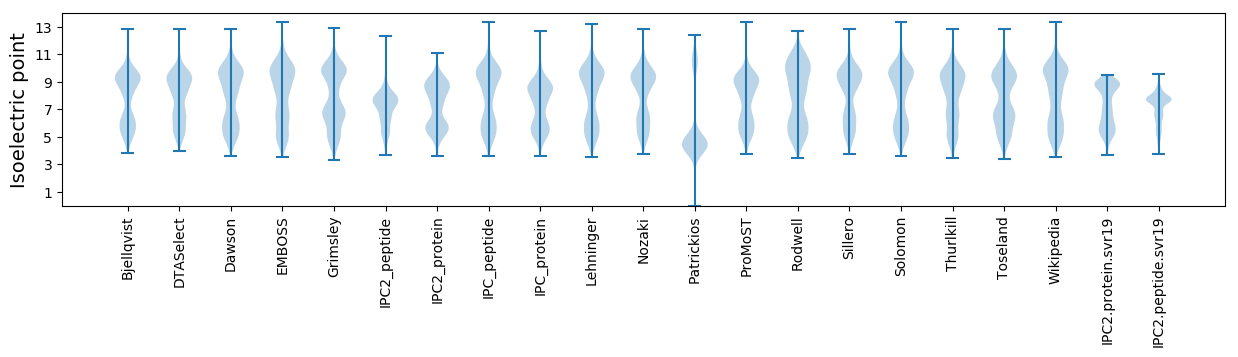
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A094JX75|A0A094JX75_9DELT PCRF domain-containing protein (Fragment) OS=Smithella sp. D17 OX=1538639 GN=KD27_05320 PE=3 SV=1
MM1 pKa = 7.73KK2 pKa = 9.08KK3 pKa = 8.39TLTNKK8 pKa = 9.48TNTKK12 pKa = 9.77RR13 pKa = 11.84KK14 pKa = 7.29KK15 pKa = 7.47THH17 pKa = 5.51GFLVRR22 pKa = 11.84MASKK26 pKa = 10.38SGRR29 pKa = 11.84QVLSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.94GRR40 pKa = 11.84KK41 pKa = 8.66RR42 pKa = 11.84LAVV45 pKa = 3.41
MM1 pKa = 7.73KK2 pKa = 9.08KK3 pKa = 8.39TLTNKK8 pKa = 9.48TNTKK12 pKa = 9.77RR13 pKa = 11.84KK14 pKa = 7.29KK15 pKa = 7.47THH17 pKa = 5.51GFLVRR22 pKa = 11.84MASKK26 pKa = 10.38SGRR29 pKa = 11.84QVLSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.94GRR40 pKa = 11.84KK41 pKa = 8.66RR42 pKa = 11.84LAVV45 pKa = 3.41
Molecular weight: 5.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
368400 |
41 |
1699 |
274.3 |
30.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.562 ± 0.063 | 1.138 ± 0.03 |
5.302 ± 0.044 | 6.45 ± 0.086 |
4.479 ± 0.06 | 6.814 ± 0.076 |
1.816 ± 0.029 | 8.217 ± 0.07 |
7.574 ± 0.079 | 9.472 ± 0.072 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.71 ± 0.03 | 4.51 ± 0.042 |
3.858 ± 0.034 | 3.2 ± 0.039 |
4.864 ± 0.058 | 6.044 ± 0.056 |
5.077 ± 0.066 | 6.543 ± 0.058 |
1.012 ± 0.028 | 3.357 ± 0.052 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
