
Acute bee paralysis virus (strain Rothamsted) (ABPV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Picornavirales; Dicistroviridae; Aparavirus; Acute bee paralysis virus
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
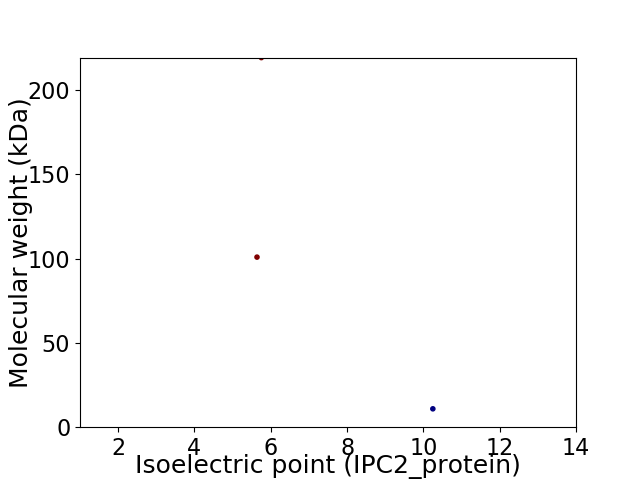
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
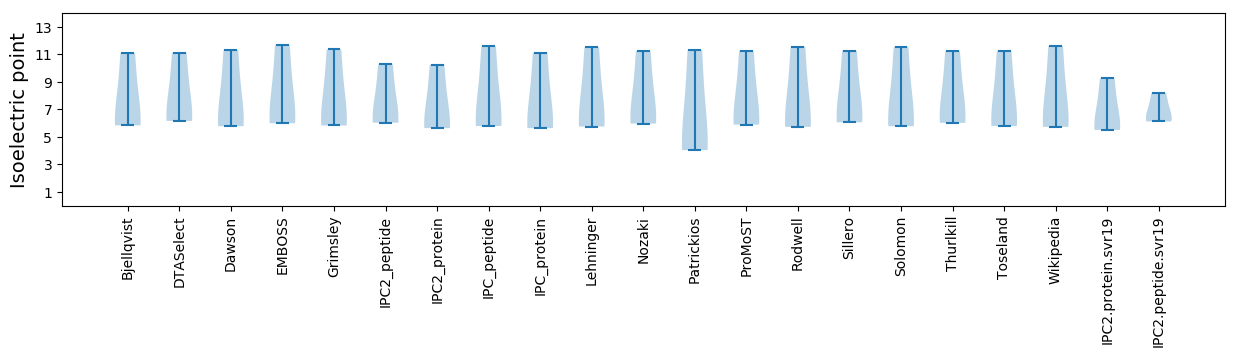
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9DSN9|POLN_ABPVR Replicase polyprotein OS=Acute bee paralysis virus (strain Rothamsted) OX=1217067 GN=ORF1 PE=4 SV=1
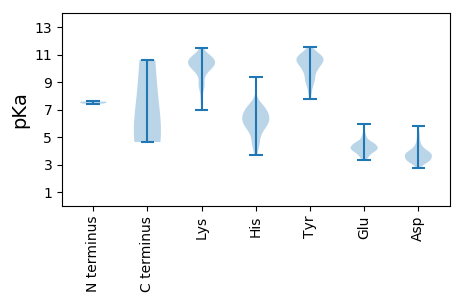
AA1 pKa = 7.58DD2 pKa = 3.66QEE4 pKa = 4.87TNTSNVHH11 pKa = 4.6NTQLASTSEE20 pKa = 4.2EE21 pKa = 3.77NSVEE25 pKa = 4.1TEE27 pKa = 4.13QITTFHH33 pKa = 7.12DD34 pKa = 3.62VEE36 pKa = 4.49TPNRR40 pKa = 11.84INTPMAQDD48 pKa = 3.47TSSARR53 pKa = 11.84SMDD56 pKa = 3.58DD57 pKa = 2.87THH59 pKa = 9.33SIIQFLQRR67 pKa = 11.84PVLIDD72 pKa = 4.19HH73 pKa = 7.02IEE75 pKa = 4.28VIAGSTADD83 pKa = 4.53DD84 pKa = 3.66NKK86 pKa = 10.59PLNRR90 pKa = 11.84YY91 pKa = 7.77VLNRR95 pKa = 11.84QNPQPFVKK103 pKa = 9.84SWTLPSVVLSAGGKK117 pKa = 7.0GQKK120 pKa = 9.3LANFKK125 pKa = 9.91YY126 pKa = 10.17LRR128 pKa = 11.84CDD130 pKa = 3.36VKK132 pKa = 11.43VKK134 pKa = 10.41IVLNANPFIAGRR146 pKa = 11.84LYY148 pKa = 10.84LAYY151 pKa = 10.2SPYY154 pKa = 10.57DD155 pKa = 3.56DD156 pKa = 4.87RR157 pKa = 11.84VDD159 pKa = 3.58PARR162 pKa = 11.84SILNTSRR169 pKa = 11.84AGVTGYY175 pKa = 9.87PGIEE179 pKa = 4.05IDD181 pKa = 3.88FQLDD185 pKa = 3.19NSVEE189 pKa = 3.97MTIPYY194 pKa = 10.63ASFQEE199 pKa = 4.96AYY201 pKa = 10.49DD202 pKa = 4.0LVTGTEE208 pKa = 4.44DD209 pKa = 3.75FVKK212 pKa = 10.63LYY214 pKa = 10.59LFTITPILSPTSTSASSKK232 pKa = 10.28VDD234 pKa = 3.17LSVYY238 pKa = 8.96MWLDD242 pKa = 3.81NISLVIPTYY251 pKa = 10.34RR252 pKa = 11.84VNTSIVPNVGTVVQTVQNMTTRR274 pKa = 11.84DD275 pKa = 3.26SEE277 pKa = 4.73TIRR280 pKa = 11.84KK281 pKa = 9.68AMVALRR287 pKa = 11.84KK288 pKa = 9.54NNKK291 pKa = 7.69STYY294 pKa = 10.09DD295 pKa = 3.73YY296 pKa = 10.72IVQALSSAVPEE307 pKa = 4.37VKK309 pKa = 10.45NVTMQINSKK318 pKa = 10.32KK319 pKa = 10.14NNSNKK324 pKa = 9.05MATPVKK330 pKa = 9.95EE331 pKa = 3.97KK332 pKa = 10.08TKK334 pKa = 10.62NIPKK338 pKa = 10.15PKK340 pKa = 9.57TEE342 pKa = 4.07NPKK345 pKa = 10.04IGPISEE351 pKa = 4.47LATGVNKK358 pKa = 9.87VANGIEE364 pKa = 4.39RR365 pKa = 11.84IPVIGEE371 pKa = 3.52MAKK374 pKa = 10.32PVTSTIKK381 pKa = 9.9WVADD385 pKa = 3.9KK386 pKa = 10.36IGSVAAIFGWSKK398 pKa = 10.33PRR400 pKa = 11.84NLEE403 pKa = 4.46QVNLYY408 pKa = 10.74QNVPGWGYY416 pKa = 11.16SLYY419 pKa = 10.75KK420 pKa = 10.82GIDD423 pKa = 3.27NSVPLAFDD431 pKa = 4.22PNNEE435 pKa = 4.23LGDD438 pKa = 3.89LRR440 pKa = 11.84DD441 pKa = 3.81VFPSGVDD448 pKa = 3.44EE449 pKa = 4.0MAIGYY454 pKa = 9.03VCGNPAVKK462 pKa = 10.1HH463 pKa = 4.5VLSWNTTDD471 pKa = 5.17KK472 pKa = 11.09VQAPISNGDD481 pKa = 3.2DD482 pKa = 3.08WGGVIPVGMPCYY494 pKa = 10.43SKK496 pKa = 10.59IIRR499 pKa = 11.84TTEE502 pKa = 3.48NDD504 pKa = 3.19TTRR507 pKa = 11.84TNTEE511 pKa = 3.43IMDD514 pKa = 4.76PAPCEE519 pKa = 4.01YY520 pKa = 9.35VCNMFSYY527 pKa = 9.67WRR529 pKa = 11.84ATMCYY534 pKa = 9.5RR535 pKa = 11.84IAIVKK540 pKa = 7.96TAFHH544 pKa = 6.72TGRR547 pKa = 11.84LGIFFGPGKK556 pKa = 10.39IPITTTKK563 pKa = 11.15DD564 pKa = 3.31NISPDD569 pKa = 3.41LTQLDD574 pKa = 4.98GIKK577 pKa = 10.57APSDD581 pKa = 3.43NNYY584 pKa = 10.67KK585 pKa = 10.78YY586 pKa = 10.65ILDD589 pKa = 3.91LTNDD593 pKa = 3.53TEE595 pKa = 4.03ITIRR599 pKa = 11.84VPFVSNKK606 pKa = 8.42MFMKK610 pKa = 9.78STGIYY615 pKa = 10.01GGNSEE620 pKa = 4.6NNWDD624 pKa = 3.7FSEE627 pKa = 4.51SFTGFLCIRR636 pKa = 11.84PITKK640 pKa = 9.49FMCPEE645 pKa = 4.03TVSNNVSIVVWKK657 pKa = 8.83WAEE660 pKa = 3.89DD661 pKa = 3.77VVVVEE666 pKa = 5.19PKK668 pKa = 10.22PLLSGPTQVFQPPVTSADD686 pKa = 3.84SINTIDD692 pKa = 5.49ASMQINLANKK702 pKa = 9.37ADD704 pKa = 3.96EE705 pKa = 4.25NVVTFFDD712 pKa = 3.84SDD714 pKa = 3.73DD715 pKa = 3.9AEE717 pKa = 4.22EE718 pKa = 4.98RR719 pKa = 11.84NMEE722 pKa = 4.4ALLKK726 pKa = 10.88GSGEE730 pKa = 4.09QIMNLRR736 pKa = 11.84SLLRR740 pKa = 11.84TFRR743 pKa = 11.84TISEE747 pKa = 4.12NWNLPPNTKK756 pKa = 8.59TAITDD761 pKa = 3.7LTDD764 pKa = 3.24VADD767 pKa = 3.96KK768 pKa = 10.84EE769 pKa = 4.51GRR771 pKa = 11.84DD772 pKa = 3.59YY773 pKa = 10.9MSYY776 pKa = 10.97LSYY779 pKa = 10.49IYY781 pKa = 10.21RR782 pKa = 11.84FYY784 pKa = 11.14RR785 pKa = 11.84GGRR788 pKa = 11.84RR789 pKa = 11.84YY790 pKa = 10.78KK791 pKa = 10.66FFNTTALKK799 pKa = 10.36QSQTCYY805 pKa = 10.22VRR807 pKa = 11.84SFLIPRR813 pKa = 11.84YY814 pKa = 8.55YY815 pKa = 10.36TADD818 pKa = 3.24NTNNDD823 pKa = 3.91GPSHH827 pKa = 5.53ITYY830 pKa = 9.6PVLNPVHH837 pKa = 6.08EE838 pKa = 4.49VEE840 pKa = 4.31VPYY843 pKa = 10.61YY844 pKa = 9.33CQYY847 pKa = 11.13RR848 pKa = 11.84KK849 pKa = 10.39LPVASTTDD857 pKa = 2.99KK858 pKa = 11.37GYY860 pKa = 10.53DD861 pKa = 3.37ASLMYY866 pKa = 10.06YY867 pKa = 10.68SNVGTNQIVARR878 pKa = 11.84AGNDD882 pKa = 3.1DD883 pKa = 4.41FTFGWLIGTPQTQGITRR900 pKa = 11.84TEE902 pKa = 4.09TKK904 pKa = 10.61
AA1 pKa = 7.58DD2 pKa = 3.66QEE4 pKa = 4.87TNTSNVHH11 pKa = 4.6NTQLASTSEE20 pKa = 4.2EE21 pKa = 3.77NSVEE25 pKa = 4.1TEE27 pKa = 4.13QITTFHH33 pKa = 7.12DD34 pKa = 3.62VEE36 pKa = 4.49TPNRR40 pKa = 11.84INTPMAQDD48 pKa = 3.47TSSARR53 pKa = 11.84SMDD56 pKa = 3.58DD57 pKa = 2.87THH59 pKa = 9.33SIIQFLQRR67 pKa = 11.84PVLIDD72 pKa = 4.19HH73 pKa = 7.02IEE75 pKa = 4.28VIAGSTADD83 pKa = 4.53DD84 pKa = 3.66NKK86 pKa = 10.59PLNRR90 pKa = 11.84YY91 pKa = 7.77VLNRR95 pKa = 11.84QNPQPFVKK103 pKa = 9.84SWTLPSVVLSAGGKK117 pKa = 7.0GQKK120 pKa = 9.3LANFKK125 pKa = 9.91YY126 pKa = 10.17LRR128 pKa = 11.84CDD130 pKa = 3.36VKK132 pKa = 11.43VKK134 pKa = 10.41IVLNANPFIAGRR146 pKa = 11.84LYY148 pKa = 10.84LAYY151 pKa = 10.2SPYY154 pKa = 10.57DD155 pKa = 3.56DD156 pKa = 4.87RR157 pKa = 11.84VDD159 pKa = 3.58PARR162 pKa = 11.84SILNTSRR169 pKa = 11.84AGVTGYY175 pKa = 9.87PGIEE179 pKa = 4.05IDD181 pKa = 3.88FQLDD185 pKa = 3.19NSVEE189 pKa = 3.97MTIPYY194 pKa = 10.63ASFQEE199 pKa = 4.96AYY201 pKa = 10.49DD202 pKa = 4.0LVTGTEE208 pKa = 4.44DD209 pKa = 3.75FVKK212 pKa = 10.63LYY214 pKa = 10.59LFTITPILSPTSTSASSKK232 pKa = 10.28VDD234 pKa = 3.17LSVYY238 pKa = 8.96MWLDD242 pKa = 3.81NISLVIPTYY251 pKa = 10.34RR252 pKa = 11.84VNTSIVPNVGTVVQTVQNMTTRR274 pKa = 11.84DD275 pKa = 3.26SEE277 pKa = 4.73TIRR280 pKa = 11.84KK281 pKa = 9.68AMVALRR287 pKa = 11.84KK288 pKa = 9.54NNKK291 pKa = 7.69STYY294 pKa = 10.09DD295 pKa = 3.73YY296 pKa = 10.72IVQALSSAVPEE307 pKa = 4.37VKK309 pKa = 10.45NVTMQINSKK318 pKa = 10.32KK319 pKa = 10.14NNSNKK324 pKa = 9.05MATPVKK330 pKa = 9.95EE331 pKa = 3.97KK332 pKa = 10.08TKK334 pKa = 10.62NIPKK338 pKa = 10.15PKK340 pKa = 9.57TEE342 pKa = 4.07NPKK345 pKa = 10.04IGPISEE351 pKa = 4.47LATGVNKK358 pKa = 9.87VANGIEE364 pKa = 4.39RR365 pKa = 11.84IPVIGEE371 pKa = 3.52MAKK374 pKa = 10.32PVTSTIKK381 pKa = 9.9WVADD385 pKa = 3.9KK386 pKa = 10.36IGSVAAIFGWSKK398 pKa = 10.33PRR400 pKa = 11.84NLEE403 pKa = 4.46QVNLYY408 pKa = 10.74QNVPGWGYY416 pKa = 11.16SLYY419 pKa = 10.75KK420 pKa = 10.82GIDD423 pKa = 3.27NSVPLAFDD431 pKa = 4.22PNNEE435 pKa = 4.23LGDD438 pKa = 3.89LRR440 pKa = 11.84DD441 pKa = 3.81VFPSGVDD448 pKa = 3.44EE449 pKa = 4.0MAIGYY454 pKa = 9.03VCGNPAVKK462 pKa = 10.1HH463 pKa = 4.5VLSWNTTDD471 pKa = 5.17KK472 pKa = 11.09VQAPISNGDD481 pKa = 3.2DD482 pKa = 3.08WGGVIPVGMPCYY494 pKa = 10.43SKK496 pKa = 10.59IIRR499 pKa = 11.84TTEE502 pKa = 3.48NDD504 pKa = 3.19TTRR507 pKa = 11.84TNTEE511 pKa = 3.43IMDD514 pKa = 4.76PAPCEE519 pKa = 4.01YY520 pKa = 9.35VCNMFSYY527 pKa = 9.67WRR529 pKa = 11.84ATMCYY534 pKa = 9.5RR535 pKa = 11.84IAIVKK540 pKa = 7.96TAFHH544 pKa = 6.72TGRR547 pKa = 11.84LGIFFGPGKK556 pKa = 10.39IPITTTKK563 pKa = 11.15DD564 pKa = 3.31NISPDD569 pKa = 3.41LTQLDD574 pKa = 4.98GIKK577 pKa = 10.57APSDD581 pKa = 3.43NNYY584 pKa = 10.67KK585 pKa = 10.78YY586 pKa = 10.65ILDD589 pKa = 3.91LTNDD593 pKa = 3.53TEE595 pKa = 4.03ITIRR599 pKa = 11.84VPFVSNKK606 pKa = 8.42MFMKK610 pKa = 9.78STGIYY615 pKa = 10.01GGNSEE620 pKa = 4.6NNWDD624 pKa = 3.7FSEE627 pKa = 4.51SFTGFLCIRR636 pKa = 11.84PITKK640 pKa = 9.49FMCPEE645 pKa = 4.03TVSNNVSIVVWKK657 pKa = 8.83WAEE660 pKa = 3.89DD661 pKa = 3.77VVVVEE666 pKa = 5.19PKK668 pKa = 10.22PLLSGPTQVFQPPVTSADD686 pKa = 3.84SINTIDD692 pKa = 5.49ASMQINLANKK702 pKa = 9.37ADD704 pKa = 3.96EE705 pKa = 4.25NVVTFFDD712 pKa = 3.84SDD714 pKa = 3.73DD715 pKa = 3.9AEE717 pKa = 4.22EE718 pKa = 4.98RR719 pKa = 11.84NMEE722 pKa = 4.4ALLKK726 pKa = 10.88GSGEE730 pKa = 4.09QIMNLRR736 pKa = 11.84SLLRR740 pKa = 11.84TFRR743 pKa = 11.84TISEE747 pKa = 4.12NWNLPPNTKK756 pKa = 8.59TAITDD761 pKa = 3.7LTDD764 pKa = 3.24VADD767 pKa = 3.96KK768 pKa = 10.84EE769 pKa = 4.51GRR771 pKa = 11.84DD772 pKa = 3.59YY773 pKa = 10.9MSYY776 pKa = 10.97LSYY779 pKa = 10.49IYY781 pKa = 10.21RR782 pKa = 11.84FYY784 pKa = 11.14RR785 pKa = 11.84GGRR788 pKa = 11.84RR789 pKa = 11.84YY790 pKa = 10.78KK791 pKa = 10.66FFNTTALKK799 pKa = 10.36QSQTCYY805 pKa = 10.22VRR807 pKa = 11.84SFLIPRR813 pKa = 11.84YY814 pKa = 8.55YY815 pKa = 10.36TADD818 pKa = 3.24NTNNDD823 pKa = 3.91GPSHH827 pKa = 5.53ITYY830 pKa = 9.6PVLNPVHH837 pKa = 6.08EE838 pKa = 4.49VEE840 pKa = 4.31VPYY843 pKa = 10.61YY844 pKa = 9.33CQYY847 pKa = 11.13RR848 pKa = 11.84KK849 pKa = 10.39LPVASTTDD857 pKa = 2.99KK858 pKa = 11.37GYY860 pKa = 10.53DD861 pKa = 3.37ASLMYY866 pKa = 10.06YY867 pKa = 10.68SNVGTNQIVARR878 pKa = 11.84AGNDD882 pKa = 3.1DD883 pKa = 4.41FTFGWLIGTPQTQGITRR900 pKa = 11.84TEE902 pKa = 4.09TKK904 pKa = 10.61
Molecular weight: 100.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9DSN8|POLS_ABPVR Structural polyprotein OS=Acute bee paralysis virus (strain Rothamsted) OX=1217067 GN=ORF2 PE=1 SV=2
PP1 pKa = 7.43IKK3 pKa = 10.59KK4 pKa = 9.97QILPTYY10 pKa = 9.89IIRR13 pKa = 11.84NSRR16 pKa = 11.84RR17 pKa = 11.84PLKK20 pKa = 9.64KK21 pKa = 8.55TQLKK25 pKa = 10.01RR26 pKa = 11.84NKK28 pKa = 9.93SPPFMMWKK36 pKa = 9.54LQIGSIPPWLKK47 pKa = 9.08TLHH50 pKa = 7.03RR51 pKa = 11.84LGAWMIRR58 pKa = 11.84TVLFSFYY65 pKa = 10.34NAPYY69 pKa = 10.62SLTTLRR75 pKa = 11.84SLLDD79 pKa = 3.56QQQMITNPSIDD90 pKa = 3.22MCC92 pKa = 5.56
PP1 pKa = 7.43IKK3 pKa = 10.59KK4 pKa = 9.97QILPTYY10 pKa = 9.89IIRR13 pKa = 11.84NSRR16 pKa = 11.84RR17 pKa = 11.84PLKK20 pKa = 9.64KK21 pKa = 8.55TQLKK25 pKa = 10.01RR26 pKa = 11.84NKK28 pKa = 9.93SPPFMMWKK36 pKa = 9.54LQIGSIPPWLKK47 pKa = 9.08TLHH50 pKa = 7.03RR51 pKa = 11.84LGAWMIRR58 pKa = 11.84TVLFSFYY65 pKa = 10.34NAPYY69 pKa = 10.62SLTTLRR75 pKa = 11.84SLLDD79 pKa = 3.56QQQMITNPSIDD90 pKa = 3.22MCC92 pKa = 5.56
Molecular weight: 10.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2902 |
92 |
1906 |
967.3 |
110.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.583 ± 0.555 | 1.654 ± 0.297 |
6.168 ± 0.612 | 6.203 ± 1.412 |
4.273 ± 0.418 | 4.962 ± 0.427 |
1.413 ± 0.272 | 7.512 ± 0.379 |
7.547 ± 0.951 | 7.788 ± 1.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.17 ± 0.432 | 6.444 ± 0.589 |
4.376 ± 1.333 | 3.377 ± 0.461 |
4.342 ± 0.481 | 6.685 ± 0.408 |
6.513 ± 1.395 | 7.478 ± 0.943 |
1.516 ± 0.257 | 3.928 ± 0.238 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
