
Anabas testudineus (Climbing perch) (Anthias testudineus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei;
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
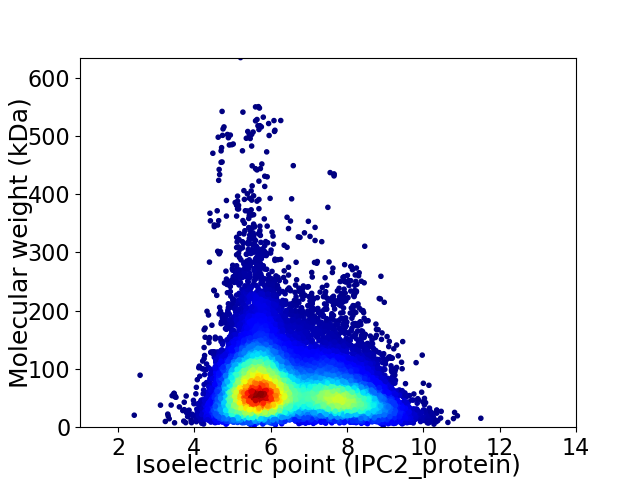
Virtual 2D-PAGE plot for 61942 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q1I070|A0A3Q1I070_ANATE RNA-binding protein 39-like OS=Anabas testudineus OX=64144 PE=4 SV=1
MM1 pKa = 7.39YY2 pKa = 10.41LEE4 pKa = 5.13NSEE7 pKa = 4.01QTDD10 pKa = 3.08ISGEE14 pKa = 4.17RR15 pKa = 11.84EE16 pKa = 3.92LSTEE20 pKa = 3.71AMLAEE25 pKa = 4.24QCQVPKK31 pKa = 9.79LTEE34 pKa = 3.96AADD37 pKa = 3.98LNDD40 pKa = 3.58ASKK43 pKa = 11.28GSAEE47 pKa = 3.86LGIRR51 pKa = 11.84VLVEE55 pKa = 3.46YY56 pKa = 10.31TEE58 pKa = 4.14QLKK61 pKa = 10.03SRR63 pKa = 11.84KK64 pKa = 8.5MDD66 pKa = 4.13FSLLCDD72 pKa = 3.84EE73 pKa = 4.62QCNMTRR79 pKa = 11.84DD80 pKa = 3.71EE81 pKa = 5.05IEE83 pKa = 3.92TFTYY87 pKa = 10.72CNGHH91 pKa = 6.42AEE93 pKa = 4.26SFEE96 pKa = 4.21QYY98 pKa = 9.95SASNGFFEE106 pKa = 4.84SDD108 pKa = 3.08QTIDD112 pKa = 2.92HH113 pKa = 6.98CEE115 pKa = 3.91TLRR118 pKa = 11.84LSQQFDD124 pKa = 3.32EE125 pKa = 4.92CAQHH129 pKa = 6.66CEE131 pKa = 4.13CFKK134 pKa = 9.87PCKK137 pKa = 10.07SSEE140 pKa = 3.83HH141 pKa = 6.43CANHH145 pKa = 6.54SLTSTCNSCCEE156 pKa = 4.01HH157 pKa = 6.74CVEE160 pKa = 4.82HH161 pKa = 6.72LQLFQQCKK169 pKa = 9.83PPDD172 pKa = 3.48QQFEE176 pKa = 4.47SFDD179 pKa = 3.68IEE181 pKa = 3.8PDD183 pKa = 3.5KK184 pKa = 11.27PVMNGDD190 pKa = 3.53NLEE193 pKa = 4.04QCEE196 pKa = 4.3MSDD199 pKa = 5.63LIPEE203 pKa = 4.43STDD206 pKa = 2.95HH207 pKa = 7.48FEE209 pKa = 4.56VLQQYY214 pKa = 9.62EE215 pKa = 4.41PTDD218 pKa = 3.76EE219 pKa = 3.97QCEE222 pKa = 4.37CFDD225 pKa = 4.92CEE227 pKa = 4.35PDD229 pKa = 3.09TSTEE233 pKa = 3.93HH234 pKa = 6.65SEE236 pKa = 3.95HH237 pKa = 8.23SEE239 pKa = 3.97MTGFTPDD246 pKa = 3.08TSDD249 pKa = 4.41SVDD252 pKa = 3.5LLDD255 pKa = 5.14YY256 pKa = 9.4GTEE259 pKa = 3.76FCEE262 pKa = 4.73YY263 pKa = 11.03DD264 pKa = 4.05DD265 pKa = 4.35IQNEE269 pKa = 4.38VATNANDD276 pKa = 3.62YY277 pKa = 11.19DD278 pKa = 4.71YY279 pKa = 11.69DD280 pKa = 6.21DD281 pKa = 6.4DD282 pKa = 7.17DD283 pKa = 6.78DD284 pKa = 6.33DD285 pKa = 5.3YY286 pKa = 12.12SEE288 pKa = 4.49NCPLEE293 pKa = 4.06QNEE296 pKa = 4.56TEE298 pKa = 4.17ASDD301 pKa = 5.36DD302 pKa = 3.68EE303 pKa = 4.6SCRR306 pKa = 11.84LSEE309 pKa = 4.55DD310 pKa = 3.73VSSSSCNTPVHH321 pKa = 6.8IYY323 pKa = 10.78YY324 pKa = 10.5DD325 pKa = 4.13DD326 pKa = 4.29NEE328 pKa = 4.57HH329 pKa = 6.19VCLFDD334 pKa = 4.7EE335 pKa = 4.53RR336 pKa = 11.84CEE338 pKa = 5.62DD339 pKa = 3.59YY340 pKa = 11.25LQTNEE345 pKa = 3.95QQAATLDD352 pKa = 3.76VSTDD356 pKa = 3.0HH357 pKa = 7.61CEE359 pKa = 3.91TCGNDD364 pKa = 3.38DD365 pKa = 3.66TEE367 pKa = 4.69TNQEE371 pKa = 4.28CEE373 pKa = 4.2PTQQCSDD380 pKa = 3.29KK381 pKa = 10.03TSEE384 pKa = 3.97FGTEE388 pKa = 4.18EE389 pKa = 4.39EE390 pKa = 4.81CSSDD394 pKa = 3.64CSSMEE399 pKa = 4.05TKK401 pKa = 10.68SFTTCPDD408 pKa = 2.76GSIPSDD414 pKa = 3.49HH415 pKa = 6.91YY416 pKa = 11.24SDD418 pKa = 4.62SSRR421 pKa = 11.84EE422 pKa = 3.86SDD424 pKa = 3.71KK425 pKa = 11.35GVQEE429 pKa = 5.27DD430 pKa = 4.03SSDD433 pKa = 4.0DD434 pKa = 3.54QTEE437 pKa = 4.24WEE439 pKa = 4.41SFEE442 pKa = 5.04EE443 pKa = 4.9DD444 pKa = 3.44EE445 pKa = 4.23QTEE448 pKa = 4.14QIKK451 pKa = 10.69INEE454 pKa = 4.32SNEE457 pKa = 4.07EE458 pKa = 4.02EE459 pKa = 4.52KK460 pKa = 9.84KK461 pKa = 9.33TPTGDD466 pKa = 3.43IVIEE470 pKa = 4.49DD471 pKa = 3.77YY472 pKa = 11.38FDD474 pKa = 4.13FFDD477 pKa = 3.52RR478 pKa = 11.84TDD480 pKa = 3.34YY481 pKa = 11.42YY482 pKa = 11.07RR483 pKa = 11.84DD484 pKa = 3.27KK485 pKa = 10.91FSQKK489 pKa = 8.27RR490 pKa = 11.84HH491 pKa = 5.95YY492 pKa = 10.19ISCFDD497 pKa = 3.9GGDD500 pKa = 3.15IHH502 pKa = 7.66DD503 pKa = 4.72CLHH506 pKa = 7.72LEE508 pKa = 4.43EE509 pKa = 5.66KK510 pKa = 9.82PQKK513 pKa = 10.4CSAKK517 pKa = 9.62NAHH520 pKa = 6.18GFEE523 pKa = 5.11KK524 pKa = 9.77ITVHH528 pKa = 6.09EE529 pKa = 4.81ADD531 pKa = 3.74VCSDD535 pKa = 3.41APEE538 pKa = 4.16EE539 pKa = 4.13ASQITSEE546 pKa = 4.14EE547 pKa = 4.24DD548 pKa = 2.81EE549 pKa = 4.45SLRR552 pKa = 11.84GDD554 pKa = 3.88SEE556 pKa = 4.64TEE558 pKa = 3.9KK559 pKa = 11.14NPEE562 pKa = 3.7DD563 pKa = 3.34WSIDD567 pKa = 3.93SEE569 pKa = 4.96SGLADD574 pKa = 4.06DD575 pKa = 6.06DD576 pKa = 3.86IEE578 pKa = 4.54EE579 pKa = 4.4IEE581 pKa = 4.74RR582 pKa = 11.84EE583 pKa = 4.28SCADD587 pKa = 3.52DD588 pKa = 3.83YY589 pKa = 12.1EE590 pKa = 4.4EE591 pKa = 6.07AEE593 pKa = 4.18EE594 pKa = 4.6DD595 pKa = 3.68ADD597 pKa = 4.82GEE599 pKa = 4.62TFDD602 pKa = 5.87EE603 pKa = 4.72EE604 pKa = 4.94ACVFDD609 pKa = 5.28GDD611 pKa = 3.89VCEE614 pKa = 4.66ICHH617 pKa = 6.14EE618 pKa = 4.52DD619 pKa = 3.37EE620 pKa = 5.73AEE622 pKa = 4.22VFSSSGNEE630 pKa = 3.73EE631 pKa = 4.47SMCAPCADD639 pKa = 4.33NISVEE644 pKa = 3.75GDD646 pKa = 3.07AYY648 pKa = 10.51EE649 pKa = 4.95DD650 pKa = 3.35EE651 pKa = 6.07GYY653 pKa = 10.63DD654 pKa = 3.87ALDD657 pKa = 3.88NEE659 pKa = 4.77SLGDD663 pKa = 3.77NTSSTDD669 pKa = 3.46QLQTTVIDD677 pKa = 3.93CKK679 pKa = 10.87KK680 pKa = 10.37DD681 pKa = 3.62NEE683 pKa = 4.52PGAEE687 pKa = 3.77DD688 pKa = 3.59TTFIEE693 pKa = 4.69CSEE696 pKa = 4.14IEE698 pKa = 4.6AYY700 pKa = 9.04WLLKK704 pKa = 10.69DD705 pKa = 3.42NEE707 pKa = 4.32YY708 pKa = 11.23NIGEE712 pKa = 4.19RR713 pKa = 11.84CEE715 pKa = 4.35SDD717 pKa = 3.38VEE719 pKa = 4.19EE720 pKa = 5.19YY721 pKa = 10.29YY722 pKa = 11.07ACQIRR727 pKa = 11.84SIQSSGKK734 pKa = 8.63QALNEE739 pKa = 3.83FMMQGQSYY747 pKa = 10.22DD748 pKa = 3.68QIIHH752 pKa = 6.58GNTNGDD758 pKa = 3.08ASRR761 pKa = 11.84SEE763 pKa = 4.17RR764 pKa = 11.84EE765 pKa = 3.84GALFPLEE772 pKa = 4.01EE773 pKa = 5.34LEE775 pKa = 4.46TEE777 pKa = 4.79GVCPEE782 pKa = 3.97EE783 pKa = 4.55SKK785 pKa = 9.9TGSFRR790 pKa = 11.84NTEE793 pKa = 3.98VGEE796 pKa = 4.01LAEE799 pKa = 5.05NLASYY804 pKa = 10.99CDD806 pKa = 3.74VEE808 pKa = 4.43NATDD812 pKa = 3.72EE813 pKa = 4.21QTQNSDD819 pKa = 3.83DD820 pKa = 3.79EE821 pKa = 4.59LKK823 pKa = 10.85EE824 pKa = 3.97VLRR827 pKa = 11.84KK828 pKa = 9.3IAPPVGIIHH837 pKa = 6.12SVVSEE842 pKa = 3.89HH843 pKa = 6.63AKK845 pKa = 9.96IAGRR849 pKa = 11.84DD850 pKa = 3.44AKK852 pKa = 10.38TEE854 pKa = 4.12EE855 pKa = 4.49IEE857 pKa = 5.45DD858 pKa = 3.96NEE860 pKa = 4.09QSSDD864 pKa = 3.82SEE866 pKa = 4.5EE867 pKa = 3.98EE868 pKa = 3.9QSDD871 pKa = 3.92NEE873 pKa = 4.22SFEE876 pKa = 4.15PCEE879 pKa = 4.51CEE881 pKa = 3.69HH882 pKa = 6.57CVPLIEE888 pKa = 4.62QVPAKK893 pKa = 10.16PLLPRR898 pKa = 11.84MKK900 pKa = 10.8SNDD903 pKa = 3.18AGKK906 pKa = 9.31ICVVIDD912 pKa = 3.71LDD914 pKa = 3.74EE915 pKa = 4.47TLVHH919 pKa = 7.06SSFKK923 pKa = 10.36PVNNADD929 pKa = 4.05FIIPVEE935 pKa = 3.92IDD937 pKa = 3.4GTVHH941 pKa = 5.33QVYY944 pKa = 9.86VLKK947 pKa = 10.52RR948 pKa = 11.84PHH950 pKa = 7.15VDD952 pKa = 2.95EE953 pKa = 4.17FLKK956 pKa = 10.99RR957 pKa = 11.84MGEE960 pKa = 3.96LFEE963 pKa = 5.41CVLFTASLAKK973 pKa = 10.69YY974 pKa = 9.38ADD976 pKa = 4.0PVSDD980 pKa = 6.19LLDD983 pKa = 3.01KK984 pKa = 10.82WGVFRR989 pKa = 11.84SRR991 pKa = 11.84LFRR994 pKa = 11.84EE995 pKa = 3.9SCVFHH1000 pKa = 6.89KK1001 pKa = 10.9GNYY1004 pKa = 9.11VKK1006 pKa = 10.65DD1007 pKa = 3.95LSRR1010 pKa = 11.84LGRR1013 pKa = 11.84DD1014 pKa = 3.3LNKK1017 pKa = 10.22VIILDD1022 pKa = 3.92NSPASYY1028 pKa = 10.25IFHH1031 pKa = 8.0PDD1033 pKa = 2.66NAVPVASWFDD1043 pKa = 3.67DD1044 pKa = 3.6MSDD1047 pKa = 3.67TEE1049 pKa = 5.5LLDD1052 pKa = 3.75LLPFFEE1058 pKa = 5.11RR1059 pKa = 11.84LSKK1062 pKa = 10.91VDD1064 pKa = 4.9DD1065 pKa = 4.3IYY1067 pKa = 11.43DD1068 pKa = 3.31ILKK1071 pKa = 8.97QQRR1074 pKa = 11.84TSSS1077 pKa = 3.31
MM1 pKa = 7.39YY2 pKa = 10.41LEE4 pKa = 5.13NSEE7 pKa = 4.01QTDD10 pKa = 3.08ISGEE14 pKa = 4.17RR15 pKa = 11.84EE16 pKa = 3.92LSTEE20 pKa = 3.71AMLAEE25 pKa = 4.24QCQVPKK31 pKa = 9.79LTEE34 pKa = 3.96AADD37 pKa = 3.98LNDD40 pKa = 3.58ASKK43 pKa = 11.28GSAEE47 pKa = 3.86LGIRR51 pKa = 11.84VLVEE55 pKa = 3.46YY56 pKa = 10.31TEE58 pKa = 4.14QLKK61 pKa = 10.03SRR63 pKa = 11.84KK64 pKa = 8.5MDD66 pKa = 4.13FSLLCDD72 pKa = 3.84EE73 pKa = 4.62QCNMTRR79 pKa = 11.84DD80 pKa = 3.71EE81 pKa = 5.05IEE83 pKa = 3.92TFTYY87 pKa = 10.72CNGHH91 pKa = 6.42AEE93 pKa = 4.26SFEE96 pKa = 4.21QYY98 pKa = 9.95SASNGFFEE106 pKa = 4.84SDD108 pKa = 3.08QTIDD112 pKa = 2.92HH113 pKa = 6.98CEE115 pKa = 3.91TLRR118 pKa = 11.84LSQQFDD124 pKa = 3.32EE125 pKa = 4.92CAQHH129 pKa = 6.66CEE131 pKa = 4.13CFKK134 pKa = 9.87PCKK137 pKa = 10.07SSEE140 pKa = 3.83HH141 pKa = 6.43CANHH145 pKa = 6.54SLTSTCNSCCEE156 pKa = 4.01HH157 pKa = 6.74CVEE160 pKa = 4.82HH161 pKa = 6.72LQLFQQCKK169 pKa = 9.83PPDD172 pKa = 3.48QQFEE176 pKa = 4.47SFDD179 pKa = 3.68IEE181 pKa = 3.8PDD183 pKa = 3.5KK184 pKa = 11.27PVMNGDD190 pKa = 3.53NLEE193 pKa = 4.04QCEE196 pKa = 4.3MSDD199 pKa = 5.63LIPEE203 pKa = 4.43STDD206 pKa = 2.95HH207 pKa = 7.48FEE209 pKa = 4.56VLQQYY214 pKa = 9.62EE215 pKa = 4.41PTDD218 pKa = 3.76EE219 pKa = 3.97QCEE222 pKa = 4.37CFDD225 pKa = 4.92CEE227 pKa = 4.35PDD229 pKa = 3.09TSTEE233 pKa = 3.93HH234 pKa = 6.65SEE236 pKa = 3.95HH237 pKa = 8.23SEE239 pKa = 3.97MTGFTPDD246 pKa = 3.08TSDD249 pKa = 4.41SVDD252 pKa = 3.5LLDD255 pKa = 5.14YY256 pKa = 9.4GTEE259 pKa = 3.76FCEE262 pKa = 4.73YY263 pKa = 11.03DD264 pKa = 4.05DD265 pKa = 4.35IQNEE269 pKa = 4.38VATNANDD276 pKa = 3.62YY277 pKa = 11.19DD278 pKa = 4.71YY279 pKa = 11.69DD280 pKa = 6.21DD281 pKa = 6.4DD282 pKa = 7.17DD283 pKa = 6.78DD284 pKa = 6.33DD285 pKa = 5.3YY286 pKa = 12.12SEE288 pKa = 4.49NCPLEE293 pKa = 4.06QNEE296 pKa = 4.56TEE298 pKa = 4.17ASDD301 pKa = 5.36DD302 pKa = 3.68EE303 pKa = 4.6SCRR306 pKa = 11.84LSEE309 pKa = 4.55DD310 pKa = 3.73VSSSSCNTPVHH321 pKa = 6.8IYY323 pKa = 10.78YY324 pKa = 10.5DD325 pKa = 4.13DD326 pKa = 4.29NEE328 pKa = 4.57HH329 pKa = 6.19VCLFDD334 pKa = 4.7EE335 pKa = 4.53RR336 pKa = 11.84CEE338 pKa = 5.62DD339 pKa = 3.59YY340 pKa = 11.25LQTNEE345 pKa = 3.95QQAATLDD352 pKa = 3.76VSTDD356 pKa = 3.0HH357 pKa = 7.61CEE359 pKa = 3.91TCGNDD364 pKa = 3.38DD365 pKa = 3.66TEE367 pKa = 4.69TNQEE371 pKa = 4.28CEE373 pKa = 4.2PTQQCSDD380 pKa = 3.29KK381 pKa = 10.03TSEE384 pKa = 3.97FGTEE388 pKa = 4.18EE389 pKa = 4.39EE390 pKa = 4.81CSSDD394 pKa = 3.64CSSMEE399 pKa = 4.05TKK401 pKa = 10.68SFTTCPDD408 pKa = 2.76GSIPSDD414 pKa = 3.49HH415 pKa = 6.91YY416 pKa = 11.24SDD418 pKa = 4.62SSRR421 pKa = 11.84EE422 pKa = 3.86SDD424 pKa = 3.71KK425 pKa = 11.35GVQEE429 pKa = 5.27DD430 pKa = 4.03SSDD433 pKa = 4.0DD434 pKa = 3.54QTEE437 pKa = 4.24WEE439 pKa = 4.41SFEE442 pKa = 5.04EE443 pKa = 4.9DD444 pKa = 3.44EE445 pKa = 4.23QTEE448 pKa = 4.14QIKK451 pKa = 10.69INEE454 pKa = 4.32SNEE457 pKa = 4.07EE458 pKa = 4.02EE459 pKa = 4.52KK460 pKa = 9.84KK461 pKa = 9.33TPTGDD466 pKa = 3.43IVIEE470 pKa = 4.49DD471 pKa = 3.77YY472 pKa = 11.38FDD474 pKa = 4.13FFDD477 pKa = 3.52RR478 pKa = 11.84TDD480 pKa = 3.34YY481 pKa = 11.42YY482 pKa = 11.07RR483 pKa = 11.84DD484 pKa = 3.27KK485 pKa = 10.91FSQKK489 pKa = 8.27RR490 pKa = 11.84HH491 pKa = 5.95YY492 pKa = 10.19ISCFDD497 pKa = 3.9GGDD500 pKa = 3.15IHH502 pKa = 7.66DD503 pKa = 4.72CLHH506 pKa = 7.72LEE508 pKa = 4.43EE509 pKa = 5.66KK510 pKa = 9.82PQKK513 pKa = 10.4CSAKK517 pKa = 9.62NAHH520 pKa = 6.18GFEE523 pKa = 5.11KK524 pKa = 9.77ITVHH528 pKa = 6.09EE529 pKa = 4.81ADD531 pKa = 3.74VCSDD535 pKa = 3.41APEE538 pKa = 4.16EE539 pKa = 4.13ASQITSEE546 pKa = 4.14EE547 pKa = 4.24DD548 pKa = 2.81EE549 pKa = 4.45SLRR552 pKa = 11.84GDD554 pKa = 3.88SEE556 pKa = 4.64TEE558 pKa = 3.9KK559 pKa = 11.14NPEE562 pKa = 3.7DD563 pKa = 3.34WSIDD567 pKa = 3.93SEE569 pKa = 4.96SGLADD574 pKa = 4.06DD575 pKa = 6.06DD576 pKa = 3.86IEE578 pKa = 4.54EE579 pKa = 4.4IEE581 pKa = 4.74RR582 pKa = 11.84EE583 pKa = 4.28SCADD587 pKa = 3.52DD588 pKa = 3.83YY589 pKa = 12.1EE590 pKa = 4.4EE591 pKa = 6.07AEE593 pKa = 4.18EE594 pKa = 4.6DD595 pKa = 3.68ADD597 pKa = 4.82GEE599 pKa = 4.62TFDD602 pKa = 5.87EE603 pKa = 4.72EE604 pKa = 4.94ACVFDD609 pKa = 5.28GDD611 pKa = 3.89VCEE614 pKa = 4.66ICHH617 pKa = 6.14EE618 pKa = 4.52DD619 pKa = 3.37EE620 pKa = 5.73AEE622 pKa = 4.22VFSSSGNEE630 pKa = 3.73EE631 pKa = 4.47SMCAPCADD639 pKa = 4.33NISVEE644 pKa = 3.75GDD646 pKa = 3.07AYY648 pKa = 10.51EE649 pKa = 4.95DD650 pKa = 3.35EE651 pKa = 6.07GYY653 pKa = 10.63DD654 pKa = 3.87ALDD657 pKa = 3.88NEE659 pKa = 4.77SLGDD663 pKa = 3.77NTSSTDD669 pKa = 3.46QLQTTVIDD677 pKa = 3.93CKK679 pKa = 10.87KK680 pKa = 10.37DD681 pKa = 3.62NEE683 pKa = 4.52PGAEE687 pKa = 3.77DD688 pKa = 3.59TTFIEE693 pKa = 4.69CSEE696 pKa = 4.14IEE698 pKa = 4.6AYY700 pKa = 9.04WLLKK704 pKa = 10.69DD705 pKa = 3.42NEE707 pKa = 4.32YY708 pKa = 11.23NIGEE712 pKa = 4.19RR713 pKa = 11.84CEE715 pKa = 4.35SDD717 pKa = 3.38VEE719 pKa = 4.19EE720 pKa = 5.19YY721 pKa = 10.29YY722 pKa = 11.07ACQIRR727 pKa = 11.84SIQSSGKK734 pKa = 8.63QALNEE739 pKa = 3.83FMMQGQSYY747 pKa = 10.22DD748 pKa = 3.68QIIHH752 pKa = 6.58GNTNGDD758 pKa = 3.08ASRR761 pKa = 11.84SEE763 pKa = 4.17RR764 pKa = 11.84EE765 pKa = 3.84GALFPLEE772 pKa = 4.01EE773 pKa = 5.34LEE775 pKa = 4.46TEE777 pKa = 4.79GVCPEE782 pKa = 3.97EE783 pKa = 4.55SKK785 pKa = 9.9TGSFRR790 pKa = 11.84NTEE793 pKa = 3.98VGEE796 pKa = 4.01LAEE799 pKa = 5.05NLASYY804 pKa = 10.99CDD806 pKa = 3.74VEE808 pKa = 4.43NATDD812 pKa = 3.72EE813 pKa = 4.21QTQNSDD819 pKa = 3.83DD820 pKa = 3.79EE821 pKa = 4.59LKK823 pKa = 10.85EE824 pKa = 3.97VLRR827 pKa = 11.84KK828 pKa = 9.3IAPPVGIIHH837 pKa = 6.12SVVSEE842 pKa = 3.89HH843 pKa = 6.63AKK845 pKa = 9.96IAGRR849 pKa = 11.84DD850 pKa = 3.44AKK852 pKa = 10.38TEE854 pKa = 4.12EE855 pKa = 4.49IEE857 pKa = 5.45DD858 pKa = 3.96NEE860 pKa = 4.09QSSDD864 pKa = 3.82SEE866 pKa = 4.5EE867 pKa = 3.98EE868 pKa = 3.9QSDD871 pKa = 3.92NEE873 pKa = 4.22SFEE876 pKa = 4.15PCEE879 pKa = 4.51CEE881 pKa = 3.69HH882 pKa = 6.57CVPLIEE888 pKa = 4.62QVPAKK893 pKa = 10.16PLLPRR898 pKa = 11.84MKK900 pKa = 10.8SNDD903 pKa = 3.18AGKK906 pKa = 9.31ICVVIDD912 pKa = 3.71LDD914 pKa = 3.74EE915 pKa = 4.47TLVHH919 pKa = 7.06SSFKK923 pKa = 10.36PVNNADD929 pKa = 4.05FIIPVEE935 pKa = 3.92IDD937 pKa = 3.4GTVHH941 pKa = 5.33QVYY944 pKa = 9.86VLKK947 pKa = 10.52RR948 pKa = 11.84PHH950 pKa = 7.15VDD952 pKa = 2.95EE953 pKa = 4.17FLKK956 pKa = 10.99RR957 pKa = 11.84MGEE960 pKa = 3.96LFEE963 pKa = 5.41CVLFTASLAKK973 pKa = 10.69YY974 pKa = 9.38ADD976 pKa = 4.0PVSDD980 pKa = 6.19LLDD983 pKa = 3.01KK984 pKa = 10.82WGVFRR989 pKa = 11.84SRR991 pKa = 11.84LFRR994 pKa = 11.84EE995 pKa = 3.9SCVFHH1000 pKa = 6.89KK1001 pKa = 10.9GNYY1004 pKa = 9.11VKK1006 pKa = 10.65DD1007 pKa = 3.95LSRR1010 pKa = 11.84LGRR1013 pKa = 11.84DD1014 pKa = 3.3LNKK1017 pKa = 10.22VIILDD1022 pKa = 3.92NSPASYY1028 pKa = 10.25IFHH1031 pKa = 8.0PDD1033 pKa = 2.66NAVPVASWFDD1043 pKa = 3.67DD1044 pKa = 3.6MSDD1047 pKa = 3.67TEE1049 pKa = 5.5LLDD1052 pKa = 3.75LLPFFEE1058 pKa = 5.11RR1059 pKa = 11.84LSKK1062 pKa = 10.91VDD1064 pKa = 4.9DD1065 pKa = 4.3IYY1067 pKa = 11.43DD1068 pKa = 3.31ILKK1071 pKa = 8.97QQRR1074 pKa = 11.84TSSS1077 pKa = 3.31
Molecular weight: 121.82 kDa
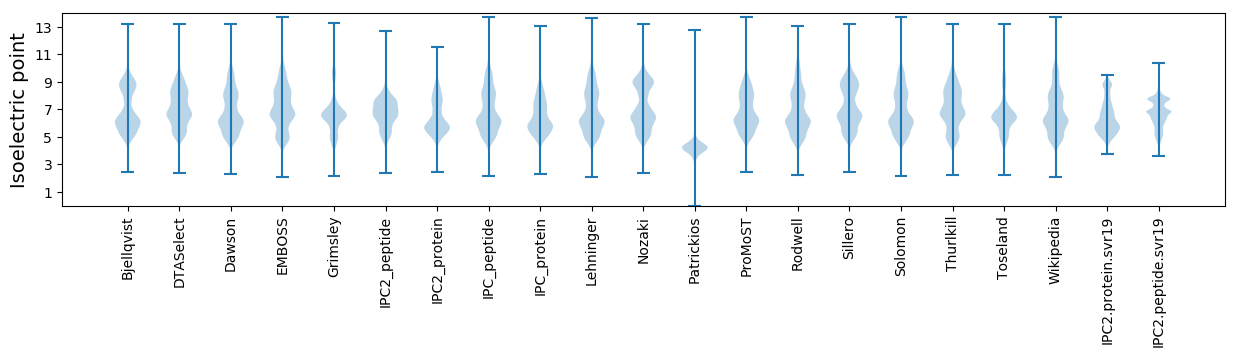
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7N6B1J7|A0A7N6B1J7_ANATE Ribosome production factor 1 homolog OS=Anabas testudineus OX=64144 GN=RPF1 PE=4 SV=1
RR1 pKa = 7.34SKK3 pKa = 9.99STALSSKK10 pKa = 9.01STALSRR16 pKa = 11.84SKK18 pKa = 9.84STALSRR24 pKa = 11.84SKK26 pKa = 10.11GTALSRR32 pKa = 11.84SKK34 pKa = 9.88STALSRR40 pKa = 11.84SKK42 pKa = 9.84STALSRR48 pKa = 11.84SKK50 pKa = 9.84STALSRR56 pKa = 11.84SKK58 pKa = 9.81STASSRR64 pKa = 11.84SKK66 pKa = 9.95GTALSRR72 pKa = 11.84SKK74 pKa = 10.16GTALSRR80 pKa = 11.84SKK82 pKa = 9.83STASSRR88 pKa = 11.84SKK90 pKa = 9.95GTALSRR96 pKa = 11.84SKK98 pKa = 9.88STALSRR104 pKa = 11.84SKK106 pKa = 10.11GTALSRR112 pKa = 11.84SKK114 pKa = 9.88STALSRR120 pKa = 11.84SKK122 pKa = 9.84STALSRR128 pKa = 11.84SKK130 pKa = 10.11GTALSRR136 pKa = 11.84SKK138 pKa = 9.88STALSRR144 pKa = 11.84SKK146 pKa = 9.92STALL150 pKa = 3.54
RR1 pKa = 7.34SKK3 pKa = 9.99STALSSKK10 pKa = 9.01STALSRR16 pKa = 11.84SKK18 pKa = 9.84STALSRR24 pKa = 11.84SKK26 pKa = 10.11GTALSRR32 pKa = 11.84SKK34 pKa = 9.88STALSRR40 pKa = 11.84SKK42 pKa = 9.84STALSRR48 pKa = 11.84SKK50 pKa = 9.84STALSRR56 pKa = 11.84SKK58 pKa = 9.81STASSRR64 pKa = 11.84SKK66 pKa = 9.95GTALSRR72 pKa = 11.84SKK74 pKa = 10.16GTALSRR80 pKa = 11.84SKK82 pKa = 9.83STASSRR88 pKa = 11.84SKK90 pKa = 9.95GTALSRR96 pKa = 11.84SKK98 pKa = 9.88STALSRR104 pKa = 11.84SKK106 pKa = 10.11GTALSRR112 pKa = 11.84SKK114 pKa = 9.88STALSRR120 pKa = 11.84SKK122 pKa = 9.84STALSRR128 pKa = 11.84SKK130 pKa = 10.11GTALSRR136 pKa = 11.84SKK138 pKa = 9.88STALSRR144 pKa = 11.84SKK146 pKa = 9.92STALL150 pKa = 3.54
Molecular weight: 15.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
42338991 |
17 |
6581 |
683.5 |
76.59 |
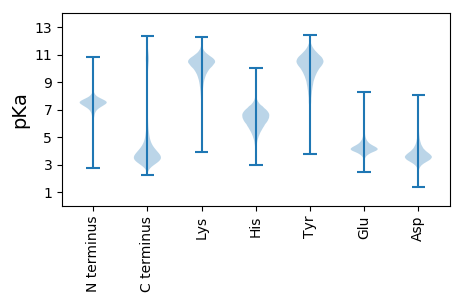
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.32 ± 0.007 | 2.316 ± 0.008 |
5.247 ± 0.006 | 6.754 ± 0.012 |
3.837 ± 0.007 | 6.182 ± 0.012 |
2.641 ± 0.004 | 4.746 ± 0.006 |
5.702 ± 0.01 | 9.743 ± 0.011 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.401 ± 0.004 | 4.016 ± 0.005 |
5.336 ± 0.01 | 4.679 ± 0.007 |
5.455 ± 0.007 | 8.291 ± 0.01 |
5.686 ± 0.007 | 6.508 ± 0.007 |
1.192 ± 0.004 | 2.948 ± 0.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
