
Enterococcus phage vB_EfaP_Ef7.2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Rountreeviridae; Sarlesvirinae; Copernicusvirus; Enterococcus virus Ef72
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
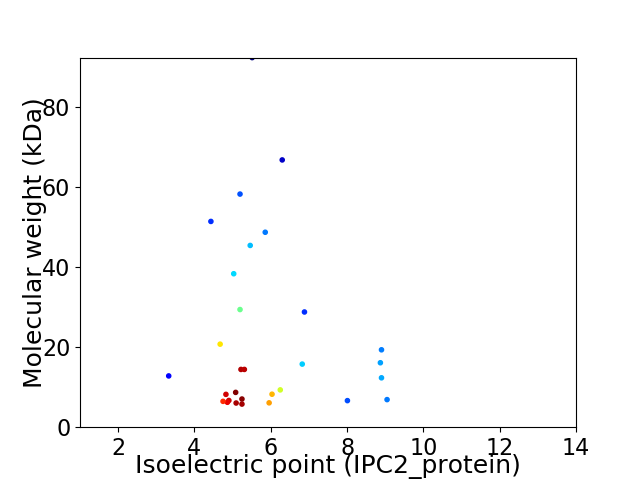
Virtual 2D-PAGE plot for 30 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6DSE4|A0A4D6DSE4_9CAUD Tail tube protein OS=Enterococcus phage vB_EfaP_Ef7.2 OX=2546618 PE=4 SV=1
MM1 pKa = 7.88PSYY4 pKa = 9.55KK5 pKa = 10.19TYY7 pKa = 8.3TTEE10 pKa = 3.54QYY12 pKa = 10.69QAFLSQPFGYY22 pKa = 10.5DD23 pKa = 2.99FGISDD28 pKa = 3.68EE29 pKa = 5.22TIAQWFMIQPGARR42 pKa = 11.84PVINSYY48 pKa = 10.97GVTKK52 pKa = 10.85ANLLSEE58 pKa = 5.04YY59 pKa = 10.08IPKK62 pKa = 10.37LKK64 pKa = 10.6QEE66 pKa = 4.42FGGSLVFLMTTVSEE80 pKa = 4.46GGGAGNWVNHH90 pKa = 5.17YY91 pKa = 10.17MSDD94 pKa = 3.44TSDD97 pKa = 2.87TGMGCMIDD105 pKa = 4.33DD106 pKa = 3.62IDD108 pKa = 4.31YY109 pKa = 10.99IKK111 pKa = 8.47TTFDD115 pKa = 2.71RR116 pKa = 11.84HH117 pKa = 5.38FPPAISAPEE126 pKa = 3.73VGGAYY131 pKa = 9.33TEE133 pKa = 4.56DD134 pKa = 3.83EE135 pKa = 4.7EE136 pKa = 4.86GLTMKK141 pKa = 10.33VYY143 pKa = 10.83NAVPDD148 pKa = 4.24GSIGSYY154 pKa = 9.63FIPSTMAGNAWIFGSQWCLANQGAAPPAVYY184 pKa = 10.15FGNPYY189 pKa = 10.29DD190 pKa = 3.84QLIDD194 pKa = 3.81IIKK197 pKa = 10.65SFGADD202 pKa = 3.34PFKK205 pKa = 10.91EE206 pKa = 4.26GSTAKK211 pKa = 10.24PNPDD215 pKa = 3.49TPKK218 pKa = 10.8GDD220 pKa = 3.97PNEE223 pKa = 4.66SGNKK227 pKa = 8.93PKK229 pKa = 10.43PKK231 pKa = 9.78PDD233 pKa = 3.01IQKK236 pKa = 10.85AIDD239 pKa = 4.27KK240 pKa = 10.73ILDD243 pKa = 4.43EE244 pKa = 4.75INKK247 pKa = 9.98ALDD250 pKa = 3.31NQTIQGSPLLSYY262 pKa = 11.17NDD264 pKa = 3.78DD265 pKa = 3.49VTIEE269 pKa = 4.01RR270 pKa = 11.84TFNNSYY276 pKa = 10.08KK277 pKa = 10.11ISYY280 pKa = 8.6TSSFKK285 pKa = 10.9KK286 pKa = 10.36KK287 pKa = 10.6LSDD290 pKa = 3.39MLNASDD296 pKa = 5.62LGLITDD302 pKa = 5.61DD303 pKa = 4.39GAQTNPPKK311 pKa = 10.21EE312 pKa = 4.25DD313 pKa = 3.69PKK315 pKa = 10.82PPSIEE320 pKa = 3.83GGKK323 pKa = 10.07DD324 pKa = 2.86SKK326 pKa = 9.77TMKK329 pKa = 10.35KK330 pKa = 10.01IYY332 pKa = 9.75DD333 pKa = 3.66WCNQNQGQAFDD344 pKa = 3.5TDD346 pKa = 3.98GYY348 pKa = 11.55YY349 pKa = 10.79GAQCVDD355 pKa = 5.11LISWINTKK363 pKa = 10.03VFGLGLDD370 pKa = 3.55TSGDD374 pKa = 3.6YY375 pKa = 11.17AKK377 pKa = 10.76NIWNNPVPSGWYY389 pKa = 9.28KK390 pKa = 11.25VNGNPNDD397 pKa = 3.97DD398 pKa = 3.77NASRR402 pKa = 11.84EE403 pKa = 3.99IWNTLPNGAIVWWTNSGAGHH423 pKa = 5.99VGIKK427 pKa = 10.48AGDD430 pKa = 3.47FATTLQQNWTSHH442 pKa = 5.43GLGGPIVLADD452 pKa = 3.69CASWMVSSGSGFLGAWVTDD471 pKa = 3.66NN472 pKa = 4.3
MM1 pKa = 7.88PSYY4 pKa = 9.55KK5 pKa = 10.19TYY7 pKa = 8.3TTEE10 pKa = 3.54QYY12 pKa = 10.69QAFLSQPFGYY22 pKa = 10.5DD23 pKa = 2.99FGISDD28 pKa = 3.68EE29 pKa = 5.22TIAQWFMIQPGARR42 pKa = 11.84PVINSYY48 pKa = 10.97GVTKK52 pKa = 10.85ANLLSEE58 pKa = 5.04YY59 pKa = 10.08IPKK62 pKa = 10.37LKK64 pKa = 10.6QEE66 pKa = 4.42FGGSLVFLMTTVSEE80 pKa = 4.46GGGAGNWVNHH90 pKa = 5.17YY91 pKa = 10.17MSDD94 pKa = 3.44TSDD97 pKa = 2.87TGMGCMIDD105 pKa = 4.33DD106 pKa = 3.62IDD108 pKa = 4.31YY109 pKa = 10.99IKK111 pKa = 8.47TTFDD115 pKa = 2.71RR116 pKa = 11.84HH117 pKa = 5.38FPPAISAPEE126 pKa = 3.73VGGAYY131 pKa = 9.33TEE133 pKa = 4.56DD134 pKa = 3.83EE135 pKa = 4.7EE136 pKa = 4.86GLTMKK141 pKa = 10.33VYY143 pKa = 10.83NAVPDD148 pKa = 4.24GSIGSYY154 pKa = 9.63FIPSTMAGNAWIFGSQWCLANQGAAPPAVYY184 pKa = 10.15FGNPYY189 pKa = 10.29DD190 pKa = 3.84QLIDD194 pKa = 3.81IIKK197 pKa = 10.65SFGADD202 pKa = 3.34PFKK205 pKa = 10.91EE206 pKa = 4.26GSTAKK211 pKa = 10.24PNPDD215 pKa = 3.49TPKK218 pKa = 10.8GDD220 pKa = 3.97PNEE223 pKa = 4.66SGNKK227 pKa = 8.93PKK229 pKa = 10.43PKK231 pKa = 9.78PDD233 pKa = 3.01IQKK236 pKa = 10.85AIDD239 pKa = 4.27KK240 pKa = 10.73ILDD243 pKa = 4.43EE244 pKa = 4.75INKK247 pKa = 9.98ALDD250 pKa = 3.31NQTIQGSPLLSYY262 pKa = 11.17NDD264 pKa = 3.78DD265 pKa = 3.49VTIEE269 pKa = 4.01RR270 pKa = 11.84TFNNSYY276 pKa = 10.08KK277 pKa = 10.11ISYY280 pKa = 8.6TSSFKK285 pKa = 10.9KK286 pKa = 10.36KK287 pKa = 10.6LSDD290 pKa = 3.39MLNASDD296 pKa = 5.62LGLITDD302 pKa = 5.61DD303 pKa = 4.39GAQTNPPKK311 pKa = 10.21EE312 pKa = 4.25DD313 pKa = 3.69PKK315 pKa = 10.82PPSIEE320 pKa = 3.83GGKK323 pKa = 10.07DD324 pKa = 2.86SKK326 pKa = 9.77TMKK329 pKa = 10.35KK330 pKa = 10.01IYY332 pKa = 9.75DD333 pKa = 3.66WCNQNQGQAFDD344 pKa = 3.5TDD346 pKa = 3.98GYY348 pKa = 11.55YY349 pKa = 10.79GAQCVDD355 pKa = 5.11LISWINTKK363 pKa = 10.03VFGLGLDD370 pKa = 3.55TSGDD374 pKa = 3.6YY375 pKa = 11.17AKK377 pKa = 10.76NIWNNPVPSGWYY389 pKa = 9.28KK390 pKa = 11.25VNGNPNDD397 pKa = 3.97DD398 pKa = 3.77NASRR402 pKa = 11.84EE403 pKa = 3.99IWNTLPNGAIVWWTNSGAGHH423 pKa = 5.99VGIKK427 pKa = 10.48AGDD430 pKa = 3.47FATTLQQNWTSHH442 pKa = 5.43GLGGPIVLADD452 pKa = 3.69CASWMVSSGSGFLGAWVTDD471 pKa = 3.66NN472 pKa = 4.3
Molecular weight: 51.43 kDa
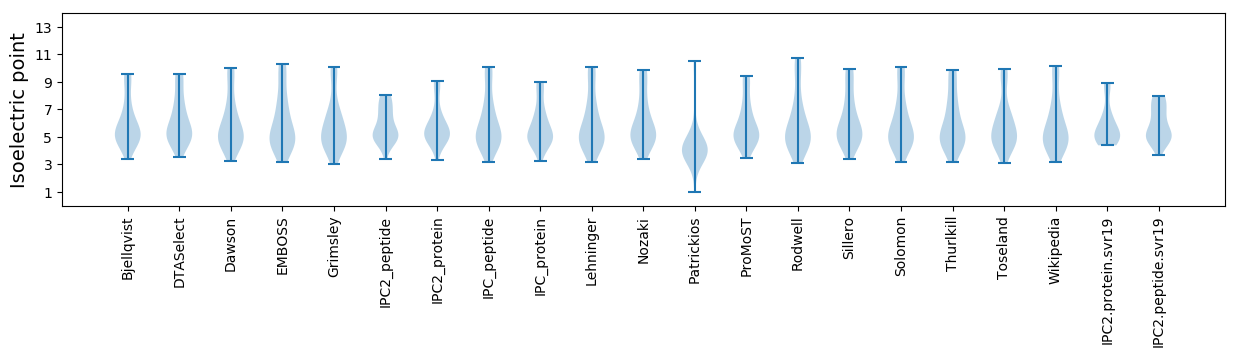
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6DSY0|A0A4D6DSY0_9CAUD Uncharacterized protein OS=Enterococcus phage vB_EfaP_Ef7.2 OX=2546618 PE=4 SV=1
MM1 pKa = 8.12RR2 pKa = 11.84YY3 pKa = 9.46KK4 pKa = 10.05QVKK7 pKa = 9.64RR8 pKa = 11.84LWEE11 pKa = 3.86LFGEE15 pKa = 4.71GEE17 pKa = 4.16YY18 pKa = 11.31GLNSFDD24 pKa = 3.57EE25 pKa = 5.0FTFNLHH31 pKa = 5.92NEE33 pKa = 4.58GIISFKK39 pKa = 10.79QYY41 pKa = 11.19LKK43 pKa = 10.66FIKK46 pKa = 9.97KK47 pKa = 9.42LKK49 pKa = 10.02KK50 pKa = 10.07KK51 pKa = 10.27EE52 pKa = 4.18GYY54 pKa = 9.12KK55 pKa = 9.94QQ56 pKa = 3.07
MM1 pKa = 8.12RR2 pKa = 11.84YY3 pKa = 9.46KK4 pKa = 10.05QVKK7 pKa = 9.64RR8 pKa = 11.84LWEE11 pKa = 3.86LFGEE15 pKa = 4.71GEE17 pKa = 4.16YY18 pKa = 11.31GLNSFDD24 pKa = 3.57EE25 pKa = 5.0FTFNLHH31 pKa = 5.92NEE33 pKa = 4.58GIISFKK39 pKa = 10.79QYY41 pKa = 11.19LKK43 pKa = 10.66FIKK46 pKa = 9.97KK47 pKa = 9.42LKK49 pKa = 10.02KK50 pKa = 10.07KK51 pKa = 10.27EE52 pKa = 4.18GYY54 pKa = 9.12KK55 pKa = 9.94QQ56 pKa = 3.07
Molecular weight: 6.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5911 |
48 |
782 |
197.0 |
22.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.211 ± 0.328 | 0.812 ± 0.143 |
6.073 ± 0.317 | 7.714 ± 0.598 |
4.838 ± 0.373 | 5.87 ± 0.538 |
1.726 ± 0.164 | 6.902 ± 0.292 |
8.239 ± 0.414 | 7.867 ± 0.468 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.876 ± 0.206 | 7.105 ± 0.406 |
3.265 ± 0.307 | 3.316 ± 0.202 |
3.282 ± 0.249 | 5.617 ± 0.36 |
6.953 ± 0.443 | 5.87 ± 0.314 |
1.286 ± 0.182 | 5.177 ± 0.285 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
