
Synechococcus phage S-CBP2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Kembevirus; Synechococcus virus SCBP2
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
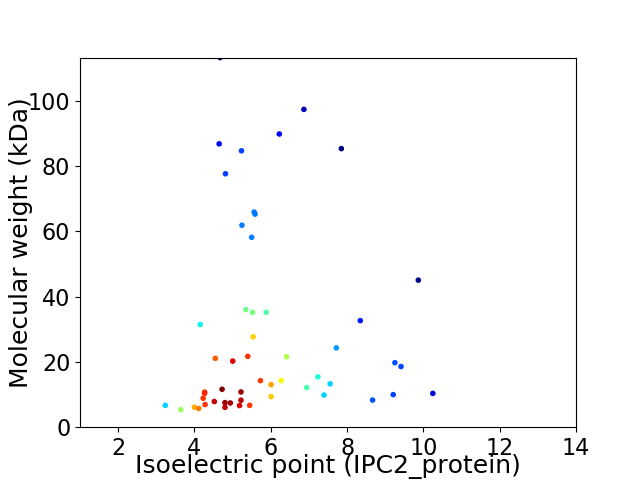
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
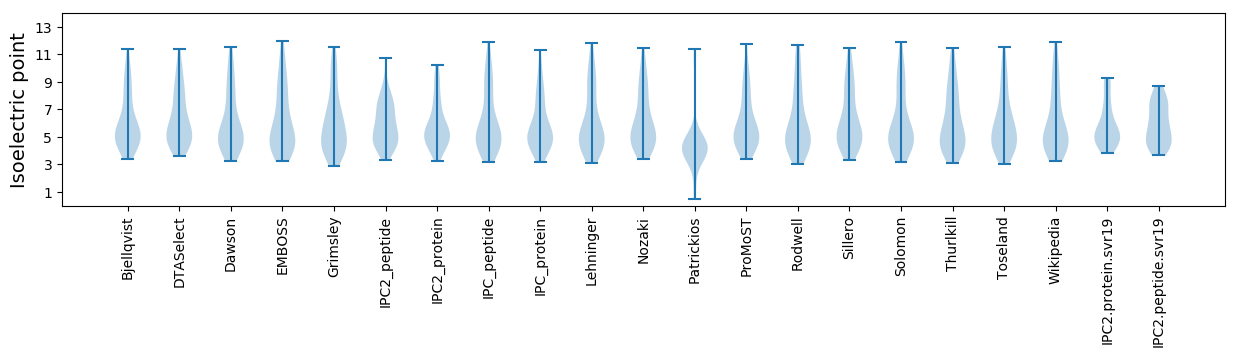
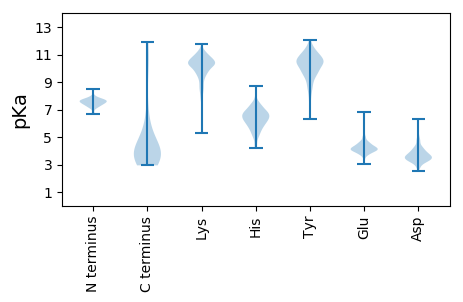
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A096VKY0|A0A096VKY0_9CAUD Uncharacterized protein OS=Synechococcus phage S-CBP2 OX=756277 GN=S-CBP2_0005 PE=4 SV=1
MM1 pKa = 7.68TYY3 pKa = 9.14LTQEE7 pKa = 4.38EE8 pKa = 4.74IEE10 pKa = 4.15NLGPEE15 pKa = 4.66EE16 pKa = 4.14YY17 pKa = 10.26CRR19 pKa = 11.84LLAYY23 pKa = 10.45GDD25 pKa = 4.06IEE27 pKa = 6.1LMDD30 pKa = 5.31DD31 pKa = 5.02DD32 pKa = 4.8EE33 pKa = 6.6FSDD36 pKa = 4.05EE37 pKa = 3.8YY38 pKa = 11.36HH39 pKa = 6.78RR40 pKa = 11.84ILRR43 pKa = 11.84HH44 pKa = 5.24FVEE47 pKa = 5.09FDD49 pKa = 2.71II50 pKa = 6.1
MM1 pKa = 7.68TYY3 pKa = 9.14LTQEE7 pKa = 4.38EE8 pKa = 4.74IEE10 pKa = 4.15NLGPEE15 pKa = 4.66EE16 pKa = 4.14YY17 pKa = 10.26CRR19 pKa = 11.84LLAYY23 pKa = 10.45GDD25 pKa = 4.06IEE27 pKa = 6.1LMDD30 pKa = 5.31DD31 pKa = 5.02DD32 pKa = 4.8EE33 pKa = 6.6FSDD36 pKa = 4.05EE37 pKa = 3.8YY38 pKa = 11.36HH39 pKa = 6.78RR40 pKa = 11.84ILRR43 pKa = 11.84HH44 pKa = 5.24FVEE47 pKa = 5.09FDD49 pKa = 2.71II50 pKa = 6.1
Molecular weight: 6.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A096VL47|A0A096VL47_9CAUD Phage_lysozyme2 domain-containing protein OS=Synechococcus phage S-CBP2 OX=756277 GN=S-CBP2_0051 PE=4 SV=1
MM1 pKa = 7.71APQNKK6 pKa = 7.78WFAASPNTDD15 pKa = 3.9YY16 pKa = 10.74IYY18 pKa = 11.08KK19 pKa = 10.35ALTDD23 pKa = 3.56GTVAKK28 pKa = 9.1LTGGRR33 pKa = 11.84IPTMNAAQAAGLVGSWLIEE52 pKa = 4.01TGRR55 pKa = 11.84KK56 pKa = 9.07DD57 pKa = 4.16LRR59 pKa = 11.84NLDD62 pKa = 3.64VVEE65 pKa = 4.72AGTGRR70 pKa = 11.84GRR72 pKa = 11.84GLSQYY77 pKa = 9.33TGVRR81 pKa = 11.84RR82 pKa = 11.84TPYY85 pKa = 10.89DD86 pKa = 3.31KK87 pKa = 10.95AVKK90 pKa = 8.93QARR93 pKa = 11.84AAGQDD98 pKa = 3.6PNSAQWQIKK107 pKa = 9.78YY108 pKa = 9.39FAQEE112 pKa = 4.05YY113 pKa = 8.7MNKK116 pKa = 10.34DD117 pKa = 3.55LIGWTKK123 pKa = 10.41VFEE126 pKa = 4.37KK127 pKa = 10.26MPKK130 pKa = 9.03NLKK133 pKa = 8.9TPGEE137 pKa = 4.04YY138 pKa = 10.39AKK140 pKa = 10.9YY141 pKa = 8.23FTGSAAEE148 pKa = 4.03GKK150 pKa = 10.42GYY152 pKa = 9.98FRR154 pKa = 11.84PGVPHH159 pKa = 6.71TDD161 pKa = 2.6RR162 pKa = 11.84RR163 pKa = 11.84MQAADD168 pKa = 3.31EE169 pKa = 4.44VFRR172 pKa = 11.84HH173 pKa = 5.04YY174 pKa = 10.77AAPNRR179 pKa = 11.84QAPANRR185 pKa = 11.84PAPAGTPAIKK195 pKa = 10.08PGPLKK200 pKa = 10.63QLMNRR205 pKa = 11.84LGIRR209 pKa = 11.84GQDD212 pKa = 2.88QGFAIDD218 pKa = 3.72KK219 pKa = 10.27LSRR222 pKa = 11.84NLGSIAAAPNAGRR235 pKa = 11.84AIFNSIAPGGNSQWKK250 pKa = 9.69SLSKK254 pKa = 10.58ADD256 pKa = 3.51KK257 pKa = 10.29QAWNTAAGQIGAQFGIAINPPTRR280 pKa = 11.84NIGLPTKK287 pKa = 10.22TNVGVNQSFGPKK299 pKa = 9.09PVNLTIRR306 pKa = 11.84GVQPGDD312 pKa = 3.34LGYY315 pKa = 8.92GTNNTRR321 pKa = 11.84VGGLPGLRR329 pKa = 11.84EE330 pKa = 3.71VAQAGIHH337 pKa = 5.73NYY339 pKa = 7.57NWKK342 pKa = 9.36PDD344 pKa = 3.99KK345 pKa = 10.86NWNSYY350 pKa = 7.32TNPHH354 pKa = 5.96YY355 pKa = 10.51NVSRR359 pKa = 11.84SSVHH363 pKa = 5.81NRR365 pKa = 11.84AGSNLRR371 pKa = 11.84GSGGLSTSSGAYY383 pKa = 9.55SGLSVGGYY391 pKa = 10.39GSGNSAYY398 pKa = 10.29RR399 pKa = 11.84GGSVSYY405 pKa = 10.93GSGSVATGTGMGSISRR421 pKa = 11.84GAGWW425 pKa = 3.21
MM1 pKa = 7.71APQNKK6 pKa = 7.78WFAASPNTDD15 pKa = 3.9YY16 pKa = 10.74IYY18 pKa = 11.08KK19 pKa = 10.35ALTDD23 pKa = 3.56GTVAKK28 pKa = 9.1LTGGRR33 pKa = 11.84IPTMNAAQAAGLVGSWLIEE52 pKa = 4.01TGRR55 pKa = 11.84KK56 pKa = 9.07DD57 pKa = 4.16LRR59 pKa = 11.84NLDD62 pKa = 3.64VVEE65 pKa = 4.72AGTGRR70 pKa = 11.84GRR72 pKa = 11.84GLSQYY77 pKa = 9.33TGVRR81 pKa = 11.84RR82 pKa = 11.84TPYY85 pKa = 10.89DD86 pKa = 3.31KK87 pKa = 10.95AVKK90 pKa = 8.93QARR93 pKa = 11.84AAGQDD98 pKa = 3.6PNSAQWQIKK107 pKa = 9.78YY108 pKa = 9.39FAQEE112 pKa = 4.05YY113 pKa = 8.7MNKK116 pKa = 10.34DD117 pKa = 3.55LIGWTKK123 pKa = 10.41VFEE126 pKa = 4.37KK127 pKa = 10.26MPKK130 pKa = 9.03NLKK133 pKa = 8.9TPGEE137 pKa = 4.04YY138 pKa = 10.39AKK140 pKa = 10.9YY141 pKa = 8.23FTGSAAEE148 pKa = 4.03GKK150 pKa = 10.42GYY152 pKa = 9.98FRR154 pKa = 11.84PGVPHH159 pKa = 6.71TDD161 pKa = 2.6RR162 pKa = 11.84RR163 pKa = 11.84MQAADD168 pKa = 3.31EE169 pKa = 4.44VFRR172 pKa = 11.84HH173 pKa = 5.04YY174 pKa = 10.77AAPNRR179 pKa = 11.84QAPANRR185 pKa = 11.84PAPAGTPAIKK195 pKa = 10.08PGPLKK200 pKa = 10.63QLMNRR205 pKa = 11.84LGIRR209 pKa = 11.84GQDD212 pKa = 2.88QGFAIDD218 pKa = 3.72KK219 pKa = 10.27LSRR222 pKa = 11.84NLGSIAAAPNAGRR235 pKa = 11.84AIFNSIAPGGNSQWKK250 pKa = 9.69SLSKK254 pKa = 10.58ADD256 pKa = 3.51KK257 pKa = 10.29QAWNTAAGQIGAQFGIAINPPTRR280 pKa = 11.84NIGLPTKK287 pKa = 10.22TNVGVNQSFGPKK299 pKa = 9.09PVNLTIRR306 pKa = 11.84GVQPGDD312 pKa = 3.34LGYY315 pKa = 8.92GTNNTRR321 pKa = 11.84VGGLPGLRR329 pKa = 11.84EE330 pKa = 3.71VAQAGIHH337 pKa = 5.73NYY339 pKa = 7.57NWKK342 pKa = 9.36PDD344 pKa = 3.99KK345 pKa = 10.86NWNSYY350 pKa = 7.32TNPHH354 pKa = 5.96YY355 pKa = 10.51NVSRR359 pKa = 11.84SSVHH363 pKa = 5.81NRR365 pKa = 11.84AGSNLRR371 pKa = 11.84GSGGLSTSSGAYY383 pKa = 9.55SGLSVGGYY391 pKa = 10.39GSGNSAYY398 pKa = 10.29RR399 pKa = 11.84GGSVSYY405 pKa = 10.93GSGSVATGTGMGSISRR421 pKa = 11.84GAGWW425 pKa = 3.21
Molecular weight: 45.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13987 |
46 |
1051 |
263.9 |
29.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.174 ± 0.396 | 0.979 ± 0.16 |
6.056 ± 0.234 | 6.256 ± 0.452 |
3.539 ± 0.202 | 7.636 ± 0.363 |
1.594 ± 0.189 | 4.619 ± 0.215 |
5.105 ± 0.323 | 8.737 ± 0.282 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.324 ± 0.227 | 4.247 ± 0.251 |
4.333 ± 0.165 | 5.119 ± 0.416 |
5.498 ± 0.22 | 6.327 ± 0.406 |
6.213 ± 0.42 | 6.728 ± 0.248 |
1.416 ± 0.14 | 3.103 ± 0.196 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
