
Fusarium poae mitovirus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.65
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
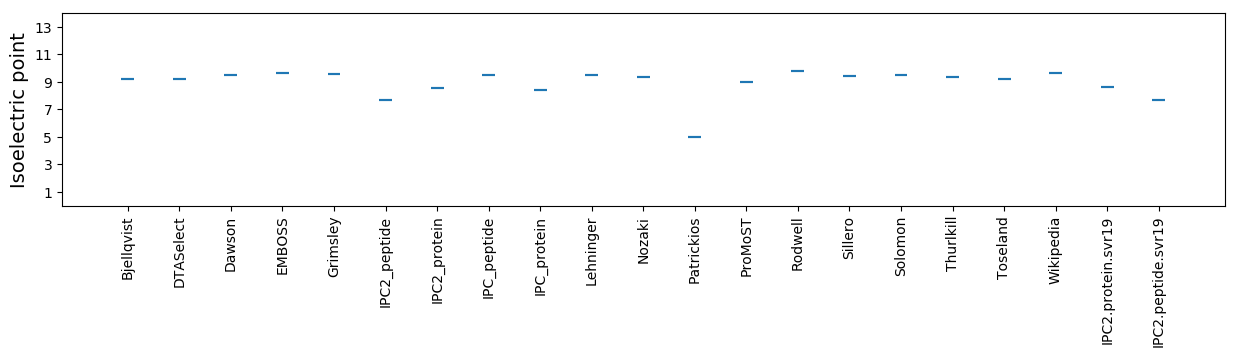
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1B4ZA33|A0A1B4ZA33_9VIRU RNA-dependent RNA polymerase OS=Fusarium poae mitovirus 4 OX=1848153 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 10.79NHH4 pKa = 6.02MKK6 pKa = 10.28LVKK9 pKa = 10.19NLGFKK14 pKa = 10.26RR15 pKa = 11.84VEE17 pKa = 4.29GGTKK21 pKa = 10.06FLNIKK26 pKa = 8.54EE27 pKa = 4.3LPRR30 pKa = 11.84FTRR33 pKa = 11.84LLIWLLASNDD43 pKa = 3.65EE44 pKa = 4.15KK45 pKa = 11.57SSLFEE50 pKa = 3.46LALRR54 pKa = 11.84IKK56 pKa = 10.57RR57 pKa = 11.84MHH59 pKa = 6.67NSNGPLFTVMYY70 pKa = 9.53LKK72 pKa = 10.45EE73 pKa = 4.16SHH75 pKa = 6.92RR76 pKa = 11.84LTMKK80 pKa = 10.87AIGGQQEE87 pKa = 4.39TCTTFPRR94 pKa = 11.84VATRR98 pKa = 11.84RR99 pKa = 11.84GLPLIVPGDD108 pKa = 3.42LRR110 pKa = 11.84VRR112 pKa = 11.84MEE114 pKa = 4.14KK115 pKa = 10.9SNFDD119 pKa = 3.71TIRR122 pKa = 11.84AVLSLLTVYY131 pKa = 10.39RR132 pKa = 11.84VINCASTLKK141 pKa = 10.54LQTITDD147 pKa = 4.09PFKK150 pKa = 10.27GTVDD154 pKa = 3.14RR155 pKa = 11.84FTSLEE160 pKa = 4.05LEE162 pKa = 4.57KK163 pKa = 10.69GFKK166 pKa = 8.9MLRR169 pKa = 11.84LKK171 pKa = 11.08GSMVLEE177 pKa = 4.13EE178 pKa = 4.57NKK180 pKa = 10.35FLIPSVKK187 pKa = 10.28SGPNYY192 pKa = 10.16KK193 pKa = 9.61IAALGATLDD202 pKa = 3.56AKK204 pKa = 10.94AFSEE208 pKa = 4.15DD209 pKa = 3.69SRR211 pKa = 11.84LLSYY215 pKa = 11.39AEE217 pKa = 4.06TVSAVTAPSLFSLLKK232 pKa = 10.83EE233 pKa = 4.38EE234 pKa = 4.38INNLGSWSSTMKK246 pKa = 10.81EE247 pKa = 4.03EE248 pKa = 4.65LLSKK252 pKa = 8.15YY253 pKa = 9.06TRR255 pKa = 11.84DD256 pKa = 3.42LKK258 pKa = 10.96LGKK261 pKa = 10.31LSEE264 pKa = 4.3KK265 pKa = 10.58KK266 pKa = 9.5EE267 pKa = 3.89AAGKK271 pKa = 10.05VRR273 pKa = 11.84VFAITDD279 pKa = 2.91VWTQSFLSPLHH290 pKa = 5.8HH291 pKa = 7.37AIFGFLKK298 pKa = 10.16RR299 pKa = 11.84IPMDD303 pKa = 3.3GTFDD307 pKa = 3.63QLKK310 pKa = 9.5PLNALLSRR318 pKa = 11.84GLKK321 pKa = 10.14NFYY324 pKa = 10.73SYY326 pKa = 11.4DD327 pKa = 3.28LSAATDD333 pKa = 3.71RR334 pKa = 11.84LPITLQEE341 pKa = 4.26QILSRR346 pKa = 11.84LFGEE350 pKa = 4.85SFAKK354 pKa = 9.68AWKK357 pKa = 9.26GLLVEE362 pKa = 4.67RR363 pKa = 11.84PWYY366 pKa = 10.25HH367 pKa = 6.7KK368 pKa = 10.26GIPYY372 pKa = 9.9LYY374 pKa = 10.64SVGQPMGALSSWGMLALTHH393 pKa = 6.49HH394 pKa = 7.01MIVQVAASRR403 pKa = 11.84VGHH406 pKa = 5.19RR407 pKa = 11.84TMFRR411 pKa = 11.84DD412 pKa = 3.61YY413 pKa = 11.56ALLGDD418 pKa = 5.54DD419 pKa = 4.01ICIADD424 pKa = 3.72SAVAKK429 pKa = 10.43SYY431 pKa = 11.13LSLMTDD437 pKa = 3.16YY438 pKa = 11.28GVDD441 pKa = 3.28INLSKK446 pKa = 10.95SLEE449 pKa = 4.04SDD451 pKa = 2.84IGVAEE456 pKa = 4.15FAKK459 pKa = 10.61RR460 pKa = 11.84LIKK463 pKa = 10.8DD464 pKa = 3.55EE465 pKa = 4.52TDD467 pKa = 3.89LSPLPPKK474 pKa = 10.72LITLLMSQFKK484 pKa = 10.5ALPTLVRR491 pKa = 11.84DD492 pKa = 3.86MIGRR496 pKa = 11.84GLSVEE501 pKa = 4.39SLLKK505 pKa = 11.0DD506 pKa = 3.24EE507 pKa = 4.88TRR509 pKa = 11.84VSRR512 pKa = 11.84PILWEE517 pKa = 4.31LIGPLGLLPSAGLSPFLGDD536 pKa = 3.78RR537 pKa = 11.84SLTKK541 pKa = 10.71DD542 pKa = 3.42EE543 pKa = 4.58LRR545 pKa = 11.84VVADD549 pKa = 3.54CVSRR553 pKa = 11.84VINRR557 pKa = 11.84WIIRR561 pKa = 11.84YY562 pKa = 8.43FYY564 pKa = 10.8QNQQTSQEE572 pKa = 4.34LIEE575 pKa = 5.01KK576 pKa = 9.95IGSLVWDD583 pKa = 4.44PSNGINRR590 pKa = 11.84DD591 pKa = 3.16TPAFHH596 pKa = 7.46HH597 pKa = 6.14YY598 pKa = 8.74MNSFIEE604 pKa = 3.99ITIEE608 pKa = 3.79EE609 pKa = 4.22NTAQPEE615 pKa = 4.47LVNFPPDD622 pKa = 3.26RR623 pKa = 11.84EE624 pKa = 4.0ASFEE628 pKa = 3.98NVYY631 pKa = 9.21TFIKK635 pKa = 10.29EE636 pKa = 4.18AMDD639 pKa = 3.98HH640 pKa = 6.71FDD642 pKa = 3.55GLAPAVPDD650 pKa = 3.28ISEE653 pKa = 4.2KK654 pKa = 10.42PRR656 pKa = 11.84VRR658 pKa = 11.84PISASSKK665 pKa = 8.02MKK667 pKa = 10.44FYY669 pKa = 11.2QEE671 pKa = 4.09LNRR674 pKa = 11.84ALQDD678 pKa = 3.05TGINFEE684 pKa = 4.38YY685 pKa = 9.76LTNNN689 pKa = 3.36
MM1 pKa = 7.47KK2 pKa = 10.79NHH4 pKa = 6.02MKK6 pKa = 10.28LVKK9 pKa = 10.19NLGFKK14 pKa = 10.26RR15 pKa = 11.84VEE17 pKa = 4.29GGTKK21 pKa = 10.06FLNIKK26 pKa = 8.54EE27 pKa = 4.3LPRR30 pKa = 11.84FTRR33 pKa = 11.84LLIWLLASNDD43 pKa = 3.65EE44 pKa = 4.15KK45 pKa = 11.57SSLFEE50 pKa = 3.46LALRR54 pKa = 11.84IKK56 pKa = 10.57RR57 pKa = 11.84MHH59 pKa = 6.67NSNGPLFTVMYY70 pKa = 9.53LKK72 pKa = 10.45EE73 pKa = 4.16SHH75 pKa = 6.92RR76 pKa = 11.84LTMKK80 pKa = 10.87AIGGQQEE87 pKa = 4.39TCTTFPRR94 pKa = 11.84VATRR98 pKa = 11.84RR99 pKa = 11.84GLPLIVPGDD108 pKa = 3.42LRR110 pKa = 11.84VRR112 pKa = 11.84MEE114 pKa = 4.14KK115 pKa = 10.9SNFDD119 pKa = 3.71TIRR122 pKa = 11.84AVLSLLTVYY131 pKa = 10.39RR132 pKa = 11.84VINCASTLKK141 pKa = 10.54LQTITDD147 pKa = 4.09PFKK150 pKa = 10.27GTVDD154 pKa = 3.14RR155 pKa = 11.84FTSLEE160 pKa = 4.05LEE162 pKa = 4.57KK163 pKa = 10.69GFKK166 pKa = 8.9MLRR169 pKa = 11.84LKK171 pKa = 11.08GSMVLEE177 pKa = 4.13EE178 pKa = 4.57NKK180 pKa = 10.35FLIPSVKK187 pKa = 10.28SGPNYY192 pKa = 10.16KK193 pKa = 9.61IAALGATLDD202 pKa = 3.56AKK204 pKa = 10.94AFSEE208 pKa = 4.15DD209 pKa = 3.69SRR211 pKa = 11.84LLSYY215 pKa = 11.39AEE217 pKa = 4.06TVSAVTAPSLFSLLKK232 pKa = 10.83EE233 pKa = 4.38EE234 pKa = 4.38INNLGSWSSTMKK246 pKa = 10.81EE247 pKa = 4.03EE248 pKa = 4.65LLSKK252 pKa = 8.15YY253 pKa = 9.06TRR255 pKa = 11.84DD256 pKa = 3.42LKK258 pKa = 10.96LGKK261 pKa = 10.31LSEE264 pKa = 4.3KK265 pKa = 10.58KK266 pKa = 9.5EE267 pKa = 3.89AAGKK271 pKa = 10.05VRR273 pKa = 11.84VFAITDD279 pKa = 2.91VWTQSFLSPLHH290 pKa = 5.8HH291 pKa = 7.37AIFGFLKK298 pKa = 10.16RR299 pKa = 11.84IPMDD303 pKa = 3.3GTFDD307 pKa = 3.63QLKK310 pKa = 9.5PLNALLSRR318 pKa = 11.84GLKK321 pKa = 10.14NFYY324 pKa = 10.73SYY326 pKa = 11.4DD327 pKa = 3.28LSAATDD333 pKa = 3.71RR334 pKa = 11.84LPITLQEE341 pKa = 4.26QILSRR346 pKa = 11.84LFGEE350 pKa = 4.85SFAKK354 pKa = 9.68AWKK357 pKa = 9.26GLLVEE362 pKa = 4.67RR363 pKa = 11.84PWYY366 pKa = 10.25HH367 pKa = 6.7KK368 pKa = 10.26GIPYY372 pKa = 9.9LYY374 pKa = 10.64SVGQPMGALSSWGMLALTHH393 pKa = 6.49HH394 pKa = 7.01MIVQVAASRR403 pKa = 11.84VGHH406 pKa = 5.19RR407 pKa = 11.84TMFRR411 pKa = 11.84DD412 pKa = 3.61YY413 pKa = 11.56ALLGDD418 pKa = 5.54DD419 pKa = 4.01ICIADD424 pKa = 3.72SAVAKK429 pKa = 10.43SYY431 pKa = 11.13LSLMTDD437 pKa = 3.16YY438 pKa = 11.28GVDD441 pKa = 3.28INLSKK446 pKa = 10.95SLEE449 pKa = 4.04SDD451 pKa = 2.84IGVAEE456 pKa = 4.15FAKK459 pKa = 10.61RR460 pKa = 11.84LIKK463 pKa = 10.8DD464 pKa = 3.55EE465 pKa = 4.52TDD467 pKa = 3.89LSPLPPKK474 pKa = 10.72LITLLMSQFKK484 pKa = 10.5ALPTLVRR491 pKa = 11.84DD492 pKa = 3.86MIGRR496 pKa = 11.84GLSVEE501 pKa = 4.39SLLKK505 pKa = 11.0DD506 pKa = 3.24EE507 pKa = 4.88TRR509 pKa = 11.84VSRR512 pKa = 11.84PILWEE517 pKa = 4.31LIGPLGLLPSAGLSPFLGDD536 pKa = 3.78RR537 pKa = 11.84SLTKK541 pKa = 10.71DD542 pKa = 3.42EE543 pKa = 4.58LRR545 pKa = 11.84VVADD549 pKa = 3.54CVSRR553 pKa = 11.84VINRR557 pKa = 11.84WIIRR561 pKa = 11.84YY562 pKa = 8.43FYY564 pKa = 10.8QNQQTSQEE572 pKa = 4.34LIEE575 pKa = 5.01KK576 pKa = 9.95IGSLVWDD583 pKa = 4.44PSNGINRR590 pKa = 11.84DD591 pKa = 3.16TPAFHH596 pKa = 7.46HH597 pKa = 6.14YY598 pKa = 8.74MNSFIEE604 pKa = 3.99ITIEE608 pKa = 3.79EE609 pKa = 4.22NTAQPEE615 pKa = 4.47LVNFPPDD622 pKa = 3.26RR623 pKa = 11.84EE624 pKa = 4.0ASFEE628 pKa = 3.98NVYY631 pKa = 9.21TFIKK635 pKa = 10.29EE636 pKa = 4.18AMDD639 pKa = 3.98HH640 pKa = 6.71FDD642 pKa = 3.55GLAPAVPDD650 pKa = 3.28ISEE653 pKa = 4.2KK654 pKa = 10.42PRR656 pKa = 11.84VRR658 pKa = 11.84PISASSKK665 pKa = 8.02MKK667 pKa = 10.44FYY669 pKa = 11.2QEE671 pKa = 4.09LNRR674 pKa = 11.84ALQDD678 pKa = 3.05TGINFEE684 pKa = 4.38YY685 pKa = 9.76LTNNN689 pKa = 3.36
Molecular weight: 77.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B4ZA33|A0A1B4ZA33_9VIRU RNA-dependent RNA polymerase OS=Fusarium poae mitovirus 4 OX=1848153 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 10.79NHH4 pKa = 6.02MKK6 pKa = 10.28LVKK9 pKa = 10.19NLGFKK14 pKa = 10.26RR15 pKa = 11.84VEE17 pKa = 4.29GGTKK21 pKa = 10.06FLNIKK26 pKa = 8.54EE27 pKa = 4.3LPRR30 pKa = 11.84FTRR33 pKa = 11.84LLIWLLASNDD43 pKa = 3.65EE44 pKa = 4.15KK45 pKa = 11.57SSLFEE50 pKa = 3.46LALRR54 pKa = 11.84IKK56 pKa = 10.57RR57 pKa = 11.84MHH59 pKa = 6.67NSNGPLFTVMYY70 pKa = 9.53LKK72 pKa = 10.45EE73 pKa = 4.16SHH75 pKa = 6.92RR76 pKa = 11.84LTMKK80 pKa = 10.87AIGGQQEE87 pKa = 4.39TCTTFPRR94 pKa = 11.84VATRR98 pKa = 11.84RR99 pKa = 11.84GLPLIVPGDD108 pKa = 3.42LRR110 pKa = 11.84VRR112 pKa = 11.84MEE114 pKa = 4.14KK115 pKa = 10.9SNFDD119 pKa = 3.71TIRR122 pKa = 11.84AVLSLLTVYY131 pKa = 10.39RR132 pKa = 11.84VINCASTLKK141 pKa = 10.54LQTITDD147 pKa = 4.09PFKK150 pKa = 10.27GTVDD154 pKa = 3.14RR155 pKa = 11.84FTSLEE160 pKa = 4.05LEE162 pKa = 4.57KK163 pKa = 10.69GFKK166 pKa = 8.9MLRR169 pKa = 11.84LKK171 pKa = 11.08GSMVLEE177 pKa = 4.13EE178 pKa = 4.57NKK180 pKa = 10.35FLIPSVKK187 pKa = 10.28SGPNYY192 pKa = 10.16KK193 pKa = 9.61IAALGATLDD202 pKa = 3.56AKK204 pKa = 10.94AFSEE208 pKa = 4.15DD209 pKa = 3.69SRR211 pKa = 11.84LLSYY215 pKa = 11.39AEE217 pKa = 4.06TVSAVTAPSLFSLLKK232 pKa = 10.83EE233 pKa = 4.38EE234 pKa = 4.38INNLGSWSSTMKK246 pKa = 10.81EE247 pKa = 4.03EE248 pKa = 4.65LLSKK252 pKa = 8.15YY253 pKa = 9.06TRR255 pKa = 11.84DD256 pKa = 3.42LKK258 pKa = 10.96LGKK261 pKa = 10.31LSEE264 pKa = 4.3KK265 pKa = 10.58KK266 pKa = 9.5EE267 pKa = 3.89AAGKK271 pKa = 10.05VRR273 pKa = 11.84VFAITDD279 pKa = 2.91VWTQSFLSPLHH290 pKa = 5.8HH291 pKa = 7.37AIFGFLKK298 pKa = 10.16RR299 pKa = 11.84IPMDD303 pKa = 3.3GTFDD307 pKa = 3.63QLKK310 pKa = 9.5PLNALLSRR318 pKa = 11.84GLKK321 pKa = 10.14NFYY324 pKa = 10.73SYY326 pKa = 11.4DD327 pKa = 3.28LSAATDD333 pKa = 3.71RR334 pKa = 11.84LPITLQEE341 pKa = 4.26QILSRR346 pKa = 11.84LFGEE350 pKa = 4.85SFAKK354 pKa = 9.68AWKK357 pKa = 9.26GLLVEE362 pKa = 4.67RR363 pKa = 11.84PWYY366 pKa = 10.25HH367 pKa = 6.7KK368 pKa = 10.26GIPYY372 pKa = 9.9LYY374 pKa = 10.64SVGQPMGALSSWGMLALTHH393 pKa = 6.49HH394 pKa = 7.01MIVQVAASRR403 pKa = 11.84VGHH406 pKa = 5.19RR407 pKa = 11.84TMFRR411 pKa = 11.84DD412 pKa = 3.61YY413 pKa = 11.56ALLGDD418 pKa = 5.54DD419 pKa = 4.01ICIADD424 pKa = 3.72SAVAKK429 pKa = 10.43SYY431 pKa = 11.13LSLMTDD437 pKa = 3.16YY438 pKa = 11.28GVDD441 pKa = 3.28INLSKK446 pKa = 10.95SLEE449 pKa = 4.04SDD451 pKa = 2.84IGVAEE456 pKa = 4.15FAKK459 pKa = 10.61RR460 pKa = 11.84LIKK463 pKa = 10.8DD464 pKa = 3.55EE465 pKa = 4.52TDD467 pKa = 3.89LSPLPPKK474 pKa = 10.72LITLLMSQFKK484 pKa = 10.5ALPTLVRR491 pKa = 11.84DD492 pKa = 3.86MIGRR496 pKa = 11.84GLSVEE501 pKa = 4.39SLLKK505 pKa = 11.0DD506 pKa = 3.24EE507 pKa = 4.88TRR509 pKa = 11.84VSRR512 pKa = 11.84PILWEE517 pKa = 4.31LIGPLGLLPSAGLSPFLGDD536 pKa = 3.78RR537 pKa = 11.84SLTKK541 pKa = 10.71DD542 pKa = 3.42EE543 pKa = 4.58LRR545 pKa = 11.84VVADD549 pKa = 3.54CVSRR553 pKa = 11.84VINRR557 pKa = 11.84WIIRR561 pKa = 11.84YY562 pKa = 8.43FYY564 pKa = 10.8QNQQTSQEE572 pKa = 4.34LIEE575 pKa = 5.01KK576 pKa = 9.95IGSLVWDD583 pKa = 4.44PSNGINRR590 pKa = 11.84DD591 pKa = 3.16TPAFHH596 pKa = 7.46HH597 pKa = 6.14YY598 pKa = 8.74MNSFIEE604 pKa = 3.99ITIEE608 pKa = 3.79EE609 pKa = 4.22NTAQPEE615 pKa = 4.47LVNFPPDD622 pKa = 3.26RR623 pKa = 11.84EE624 pKa = 4.0ASFEE628 pKa = 3.98NVYY631 pKa = 9.21TFIKK635 pKa = 10.29EE636 pKa = 4.18AMDD639 pKa = 3.98HH640 pKa = 6.71FDD642 pKa = 3.55GLAPAVPDD650 pKa = 3.28ISEE653 pKa = 4.2KK654 pKa = 10.42PRR656 pKa = 11.84VRR658 pKa = 11.84PISASSKK665 pKa = 8.02MKK667 pKa = 10.44FYY669 pKa = 11.2QEE671 pKa = 4.09LNRR674 pKa = 11.84ALQDD678 pKa = 3.05TGINFEE684 pKa = 4.38YY685 pKa = 9.76LTNNN689 pKa = 3.36
MM1 pKa = 7.47KK2 pKa = 10.79NHH4 pKa = 6.02MKK6 pKa = 10.28LVKK9 pKa = 10.19NLGFKK14 pKa = 10.26RR15 pKa = 11.84VEE17 pKa = 4.29GGTKK21 pKa = 10.06FLNIKK26 pKa = 8.54EE27 pKa = 4.3LPRR30 pKa = 11.84FTRR33 pKa = 11.84LLIWLLASNDD43 pKa = 3.65EE44 pKa = 4.15KK45 pKa = 11.57SSLFEE50 pKa = 3.46LALRR54 pKa = 11.84IKK56 pKa = 10.57RR57 pKa = 11.84MHH59 pKa = 6.67NSNGPLFTVMYY70 pKa = 9.53LKK72 pKa = 10.45EE73 pKa = 4.16SHH75 pKa = 6.92RR76 pKa = 11.84LTMKK80 pKa = 10.87AIGGQQEE87 pKa = 4.39TCTTFPRR94 pKa = 11.84VATRR98 pKa = 11.84RR99 pKa = 11.84GLPLIVPGDD108 pKa = 3.42LRR110 pKa = 11.84VRR112 pKa = 11.84MEE114 pKa = 4.14KK115 pKa = 10.9SNFDD119 pKa = 3.71TIRR122 pKa = 11.84AVLSLLTVYY131 pKa = 10.39RR132 pKa = 11.84VINCASTLKK141 pKa = 10.54LQTITDD147 pKa = 4.09PFKK150 pKa = 10.27GTVDD154 pKa = 3.14RR155 pKa = 11.84FTSLEE160 pKa = 4.05LEE162 pKa = 4.57KK163 pKa = 10.69GFKK166 pKa = 8.9MLRR169 pKa = 11.84LKK171 pKa = 11.08GSMVLEE177 pKa = 4.13EE178 pKa = 4.57NKK180 pKa = 10.35FLIPSVKK187 pKa = 10.28SGPNYY192 pKa = 10.16KK193 pKa = 9.61IAALGATLDD202 pKa = 3.56AKK204 pKa = 10.94AFSEE208 pKa = 4.15DD209 pKa = 3.69SRR211 pKa = 11.84LLSYY215 pKa = 11.39AEE217 pKa = 4.06TVSAVTAPSLFSLLKK232 pKa = 10.83EE233 pKa = 4.38EE234 pKa = 4.38INNLGSWSSTMKK246 pKa = 10.81EE247 pKa = 4.03EE248 pKa = 4.65LLSKK252 pKa = 8.15YY253 pKa = 9.06TRR255 pKa = 11.84DD256 pKa = 3.42LKK258 pKa = 10.96LGKK261 pKa = 10.31LSEE264 pKa = 4.3KK265 pKa = 10.58KK266 pKa = 9.5EE267 pKa = 3.89AAGKK271 pKa = 10.05VRR273 pKa = 11.84VFAITDD279 pKa = 2.91VWTQSFLSPLHH290 pKa = 5.8HH291 pKa = 7.37AIFGFLKK298 pKa = 10.16RR299 pKa = 11.84IPMDD303 pKa = 3.3GTFDD307 pKa = 3.63QLKK310 pKa = 9.5PLNALLSRR318 pKa = 11.84GLKK321 pKa = 10.14NFYY324 pKa = 10.73SYY326 pKa = 11.4DD327 pKa = 3.28LSAATDD333 pKa = 3.71RR334 pKa = 11.84LPITLQEE341 pKa = 4.26QILSRR346 pKa = 11.84LFGEE350 pKa = 4.85SFAKK354 pKa = 9.68AWKK357 pKa = 9.26GLLVEE362 pKa = 4.67RR363 pKa = 11.84PWYY366 pKa = 10.25HH367 pKa = 6.7KK368 pKa = 10.26GIPYY372 pKa = 9.9LYY374 pKa = 10.64SVGQPMGALSSWGMLALTHH393 pKa = 6.49HH394 pKa = 7.01MIVQVAASRR403 pKa = 11.84VGHH406 pKa = 5.19RR407 pKa = 11.84TMFRR411 pKa = 11.84DD412 pKa = 3.61YY413 pKa = 11.56ALLGDD418 pKa = 5.54DD419 pKa = 4.01ICIADD424 pKa = 3.72SAVAKK429 pKa = 10.43SYY431 pKa = 11.13LSLMTDD437 pKa = 3.16YY438 pKa = 11.28GVDD441 pKa = 3.28INLSKK446 pKa = 10.95SLEE449 pKa = 4.04SDD451 pKa = 2.84IGVAEE456 pKa = 4.15FAKK459 pKa = 10.61RR460 pKa = 11.84LIKK463 pKa = 10.8DD464 pKa = 3.55EE465 pKa = 4.52TDD467 pKa = 3.89LSPLPPKK474 pKa = 10.72LITLLMSQFKK484 pKa = 10.5ALPTLVRR491 pKa = 11.84DD492 pKa = 3.86MIGRR496 pKa = 11.84GLSVEE501 pKa = 4.39SLLKK505 pKa = 11.0DD506 pKa = 3.24EE507 pKa = 4.88TRR509 pKa = 11.84VSRR512 pKa = 11.84PILWEE517 pKa = 4.31LIGPLGLLPSAGLSPFLGDD536 pKa = 3.78RR537 pKa = 11.84SLTKK541 pKa = 10.71DD542 pKa = 3.42EE543 pKa = 4.58LRR545 pKa = 11.84VVADD549 pKa = 3.54CVSRR553 pKa = 11.84VINRR557 pKa = 11.84WIIRR561 pKa = 11.84YY562 pKa = 8.43FYY564 pKa = 10.8QNQQTSQEE572 pKa = 4.34LIEE575 pKa = 5.01KK576 pKa = 9.95IGSLVWDD583 pKa = 4.44PSNGINRR590 pKa = 11.84DD591 pKa = 3.16TPAFHH596 pKa = 7.46HH597 pKa = 6.14YY598 pKa = 8.74MNSFIEE604 pKa = 3.99ITIEE608 pKa = 3.79EE609 pKa = 4.22NTAQPEE615 pKa = 4.47LVNFPPDD622 pKa = 3.26RR623 pKa = 11.84EE624 pKa = 4.0ASFEE628 pKa = 3.98NVYY631 pKa = 9.21TFIKK635 pKa = 10.29EE636 pKa = 4.18AMDD639 pKa = 3.98HH640 pKa = 6.71FDD642 pKa = 3.55GLAPAVPDD650 pKa = 3.28ISEE653 pKa = 4.2KK654 pKa = 10.42PRR656 pKa = 11.84VRR658 pKa = 11.84PISASSKK665 pKa = 8.02MKK667 pKa = 10.44FYY669 pKa = 11.2QEE671 pKa = 4.09LNRR674 pKa = 11.84ALQDD678 pKa = 3.05TGINFEE684 pKa = 4.38YY685 pKa = 9.76LTNNN689 pKa = 3.36
Molecular weight: 77.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
689 |
689 |
689 |
689.0 |
77.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.386 ± 0.0 | 0.581 ± 0.0 |
4.935 ± 0.0 | 5.951 ± 0.0 |
4.935 ± 0.0 | 5.66 ± 0.0 |
1.742 ± 0.0 | 5.806 ± 0.0 |
6.821 ± 0.0 | 13.498 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.903 ± 0.0 | 3.919 ± 0.0 |
4.79 ± 0.0 | 2.467 ± 0.0 |
5.951 ± 0.0 | 8.418 ± 0.0 |
5.806 ± 0.0 | 5.37 ± 0.0 |
1.306 ± 0.0 | 2.758 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
