
Veillonella sp. AS16
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Negativicutes; Veillonellales; Veillonellaceae; Veillonella; unclassified Veillonella
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
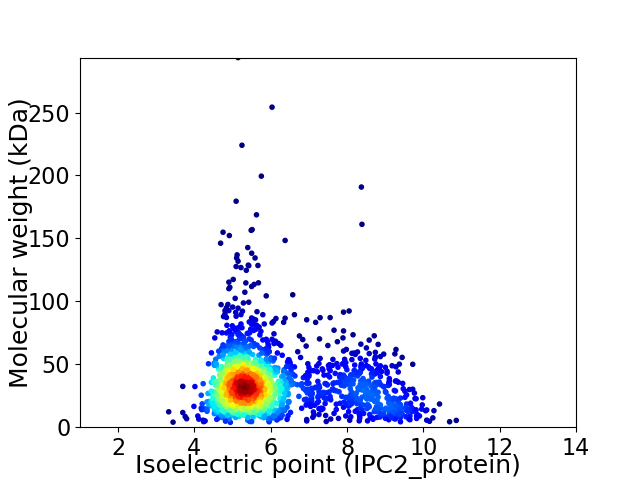
Virtual 2D-PAGE plot for 1736 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W3Y561|W3Y561_9FIRM 50S ribosomal protein L21 OS=Veillonella sp. AS16 OX=936589 GN=rplU PE=3 SV=1
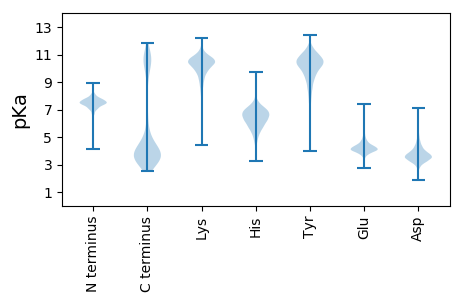
MM1 pKa = 7.52TEE3 pKa = 3.6IQALLLLKK11 pKa = 10.73ASLEE15 pKa = 4.24SADD18 pKa = 3.25IHH20 pKa = 5.8GVEE23 pKa = 5.41SILSDD28 pKa = 3.35DD29 pKa = 4.49CEE31 pKa = 4.38YY32 pKa = 10.69MSTGRR37 pKa = 11.84GIIGRR42 pKa = 11.84NRR44 pKa = 11.84SEE46 pKa = 4.24TIDD49 pKa = 3.76FLSSMMEE56 pKa = 4.09SIKK59 pKa = 10.7ADD61 pKa = 3.37RR62 pKa = 11.84VPVNCNVMHH71 pKa = 6.23ITDD74 pKa = 3.77VYY76 pKa = 11.29EE77 pKa = 4.36EE78 pKa = 4.52GALFATGRR86 pKa = 11.84HH87 pKa = 5.28GLSVSYY93 pKa = 10.5EE94 pKa = 4.03SGDD97 pKa = 3.5TYY99 pKa = 11.61VYY101 pKa = 10.58MLFIDD106 pKa = 4.09VNDD109 pKa = 3.73EE110 pKa = 4.07GLVDD114 pKa = 5.33RR115 pKa = 11.84IVSSQEE121 pKa = 3.84NYY123 pKa = 10.59SIAYY127 pKa = 7.48DD128 pKa = 3.75TLRR131 pKa = 11.84LNDD134 pKa = 4.96DD135 pKa = 3.51EE136 pKa = 5.07PEE138 pKa = 4.31YY139 pKa = 11.37AFTTAPTTVKK149 pKa = 10.7DD150 pKa = 3.72WITALSIWLEE160 pKa = 4.04TDD162 pKa = 3.02NVDD165 pKa = 3.11IDD167 pKa = 3.75DD168 pKa = 4.73FYY170 pKa = 11.67RR171 pKa = 11.84YY172 pKa = 8.99IDD174 pKa = 3.51EE175 pKa = 4.23DD176 pKa = 3.93TRR178 pKa = 11.84VVFQKK183 pKa = 11.05GDD185 pKa = 3.45AEE187 pKa = 5.22SITLEE192 pKa = 4.12NGLDD196 pKa = 3.43VEE198 pKa = 5.73DD199 pKa = 5.74YY200 pKa = 10.97FDD202 pKa = 4.19QLMNQHH208 pKa = 6.37FDD210 pKa = 3.52AVPHH214 pKa = 7.17IIRR217 pKa = 11.84DD218 pKa = 3.73DD219 pKa = 4.22DD220 pKa = 4.8DD221 pKa = 5.26NLILTYY227 pKa = 11.14GPMSLIATLNEE238 pKa = 4.35DD239 pKa = 3.35GALALISIYY248 pKa = 10.26IDD250 pKa = 3.02IRR252 pKa = 11.84DD253 pKa = 3.77EE254 pKa = 4.24DD255 pKa = 3.95VSDD258 pKa = 3.36EE259 pKa = 4.1VGYY262 pKa = 10.4IEE264 pKa = 5.47EE265 pKa = 5.98DD266 pKa = 3.01ITEE269 pKa = 4.66IIYY272 pKa = 10.73DD273 pKa = 4.15DD274 pKa = 3.87EE275 pKa = 7.39DD276 pKa = 3.65ITEE279 pKa = 4.13VAHH282 pKa = 7.89DD283 pKa = 4.3DD284 pKa = 4.11FEE286 pKa = 4.57
MM1 pKa = 7.52TEE3 pKa = 3.6IQALLLLKK11 pKa = 10.73ASLEE15 pKa = 4.24SADD18 pKa = 3.25IHH20 pKa = 5.8GVEE23 pKa = 5.41SILSDD28 pKa = 3.35DD29 pKa = 4.49CEE31 pKa = 4.38YY32 pKa = 10.69MSTGRR37 pKa = 11.84GIIGRR42 pKa = 11.84NRR44 pKa = 11.84SEE46 pKa = 4.24TIDD49 pKa = 3.76FLSSMMEE56 pKa = 4.09SIKK59 pKa = 10.7ADD61 pKa = 3.37RR62 pKa = 11.84VPVNCNVMHH71 pKa = 6.23ITDD74 pKa = 3.77VYY76 pKa = 11.29EE77 pKa = 4.36EE78 pKa = 4.52GALFATGRR86 pKa = 11.84HH87 pKa = 5.28GLSVSYY93 pKa = 10.5EE94 pKa = 4.03SGDD97 pKa = 3.5TYY99 pKa = 11.61VYY101 pKa = 10.58MLFIDD106 pKa = 4.09VNDD109 pKa = 3.73EE110 pKa = 4.07GLVDD114 pKa = 5.33RR115 pKa = 11.84IVSSQEE121 pKa = 3.84NYY123 pKa = 10.59SIAYY127 pKa = 7.48DD128 pKa = 3.75TLRR131 pKa = 11.84LNDD134 pKa = 4.96DD135 pKa = 3.51EE136 pKa = 5.07PEE138 pKa = 4.31YY139 pKa = 11.37AFTTAPTTVKK149 pKa = 10.7DD150 pKa = 3.72WITALSIWLEE160 pKa = 4.04TDD162 pKa = 3.02NVDD165 pKa = 3.11IDD167 pKa = 3.75DD168 pKa = 4.73FYY170 pKa = 11.67RR171 pKa = 11.84YY172 pKa = 8.99IDD174 pKa = 3.51EE175 pKa = 4.23DD176 pKa = 3.93TRR178 pKa = 11.84VVFQKK183 pKa = 11.05GDD185 pKa = 3.45AEE187 pKa = 5.22SITLEE192 pKa = 4.12NGLDD196 pKa = 3.43VEE198 pKa = 5.73DD199 pKa = 5.74YY200 pKa = 10.97FDD202 pKa = 4.19QLMNQHH208 pKa = 6.37FDD210 pKa = 3.52AVPHH214 pKa = 7.17IIRR217 pKa = 11.84DD218 pKa = 3.73DD219 pKa = 4.22DD220 pKa = 4.8DD221 pKa = 5.26NLILTYY227 pKa = 11.14GPMSLIATLNEE238 pKa = 4.35DD239 pKa = 3.35GALALISIYY248 pKa = 10.26IDD250 pKa = 3.02IRR252 pKa = 11.84DD253 pKa = 3.77EE254 pKa = 4.24DD255 pKa = 3.95VSDD258 pKa = 3.36EE259 pKa = 4.1VGYY262 pKa = 10.4IEE264 pKa = 5.47EE265 pKa = 5.98DD266 pKa = 3.01ITEE269 pKa = 4.66IIYY272 pKa = 10.73DD273 pKa = 4.15DD274 pKa = 3.87EE275 pKa = 7.39DD276 pKa = 3.65ITEE279 pKa = 4.13VAHH282 pKa = 7.89DD283 pKa = 4.3DD284 pKa = 4.11FEE286 pKa = 4.57
Molecular weight: 32.42 kDa
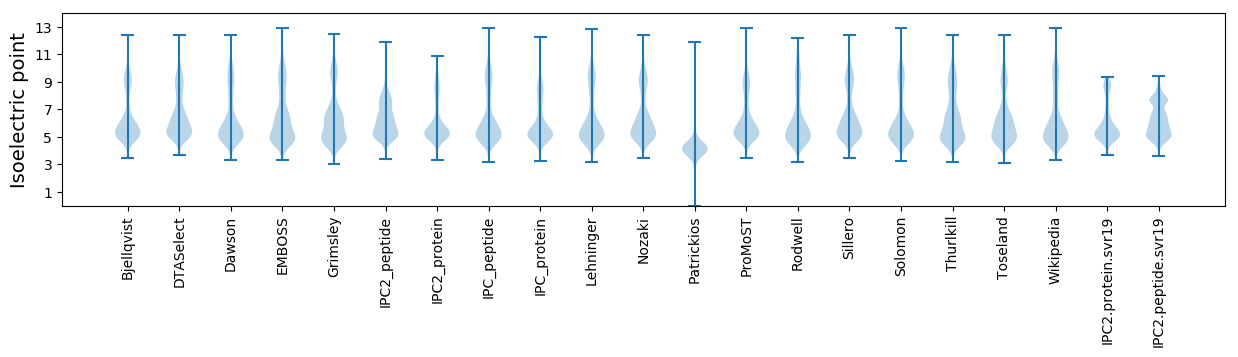
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W3Y1K6|W3Y1K6_9FIRM BMC domain protein OS=Veillonella sp. AS16 OX=936589 GN=HMPREF1521_0856 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.31QPNTLWRR12 pKa = 11.84KK13 pKa = 6.72RR14 pKa = 11.84THH16 pKa = 6.25GFRR19 pKa = 11.84EE20 pKa = 3.78RR21 pKa = 11.84MKK23 pKa = 10.34TIGGRR28 pKa = 11.84LVLKK32 pKa = 10.36RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.31QPNTLWRR12 pKa = 11.84KK13 pKa = 6.72RR14 pKa = 11.84THH16 pKa = 6.25GFRR19 pKa = 11.84EE20 pKa = 3.78RR21 pKa = 11.84MKK23 pKa = 10.34TIGGRR28 pKa = 11.84LVLKK32 pKa = 10.36RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
572448 |
34 |
2862 |
329.8 |
36.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.23 ± 0.066 | 1.035 ± 0.022 |
5.876 ± 0.047 | 6.316 ± 0.068 |
3.779 ± 0.045 | 7.589 ± 0.069 |
2.235 ± 0.032 | 7.486 ± 0.063 |
5.851 ± 0.056 | 9.002 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.008 ± 0.031 | 4.293 ± 0.063 |
3.699 ± 0.036 | 3.353 ± 0.037 |
4.526 ± 0.05 | 5.683 ± 0.043 |
5.798 ± 0.061 | 7.747 ± 0.05 |
0.9 ± 0.022 | 3.594 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
