
Lake Sarah-associated circular virus-42
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.68
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A140AQQ8|A0A140AQQ8_9VIRU Coat protein OS=Lake Sarah-associated circular virus-42 OX=1685771 PE=4 SV=1
MM1 pKa = 7.42SRR3 pKa = 11.84GKK5 pKa = 9.38SWCFTIHH12 pKa = 6.6SRR14 pKa = 11.84GEE16 pKa = 3.95GDD18 pKa = 3.41DD19 pKa = 4.79CDD21 pKa = 4.08WLLHH25 pKa = 6.3PIGTAEE31 pKa = 4.35PPSAFWDD38 pKa = 3.81DD39 pKa = 5.84DD40 pKa = 3.96PFEE43 pKa = 4.51WGCQFMIFQMEE54 pKa = 4.53RR55 pKa = 11.84CPEE58 pKa = 3.95TGKK61 pKa = 10.25LHH63 pKa = 6.41LQGALQLDD71 pKa = 3.82KK72 pKa = 10.83QQRR75 pKa = 11.84LSFMKK80 pKa = 10.32KK81 pKa = 7.85LHH83 pKa = 6.07KK84 pKa = 8.13TAHH87 pKa = 5.58WEE89 pKa = 4.07VMKK92 pKa = 10.98GNWTQSIEE100 pKa = 4.17YY101 pKa = 9.49CSKK104 pKa = 11.16SEE106 pKa = 4.15TKK108 pKa = 10.89VNGPWEE114 pKa = 4.34AGDD117 pKa = 3.8RR118 pKa = 11.84PKK120 pKa = 10.32MGQRR124 pKa = 11.84TDD126 pKa = 3.38LEE128 pKa = 4.9TIGKK132 pKa = 7.56MVKK135 pKa = 9.52EE136 pKa = 3.98NKK138 pKa = 9.25TNLEE142 pKa = 4.08LVDD145 pKa = 3.65TLGAGVSKK153 pKa = 9.79FQKK156 pKa = 9.99HH157 pKa = 4.44ISFLRR162 pKa = 11.84FTYY165 pKa = 9.9SEE167 pKa = 4.3KK168 pKa = 11.05DD169 pKa = 3.03SDD171 pKa = 3.85RR172 pKa = 11.84QATGVKK178 pKa = 10.15VIVLYY183 pKa = 11.07GPTGTGKK190 pKa = 8.99TYY192 pKa = 10.95AAVNSLTGGRR202 pKa = 11.84AYY204 pKa = 10.49HH205 pKa = 6.72ILNAPSQKK213 pKa = 9.11ATKK216 pKa = 9.64LWFDD220 pKa = 3.52GYY222 pKa = 10.92EE223 pKa = 3.98NQKK226 pKa = 10.26VLVIDD231 pKa = 4.82DD232 pKa = 4.01FDD234 pKa = 6.0GSVEE238 pKa = 3.85FRR240 pKa = 11.84YY241 pKa = 9.93LLRR244 pKa = 11.84MIDD247 pKa = 3.69VYY249 pKa = 11.21KK250 pKa = 10.75FPAEE254 pKa = 4.31VKK256 pKa = 10.45GGMVWGVWDD265 pKa = 3.82TVIITSNVHH274 pKa = 5.02PASWYY279 pKa = 8.69TGVDD283 pKa = 3.22TSPLKK288 pKa = 10.61RR289 pKa = 11.84RR290 pKa = 11.84IAEE293 pKa = 3.62IRR295 pKa = 11.84LCEE298 pKa = 3.93NQGTYY303 pKa = 11.17KK304 pKa = 9.94MISWEE309 pKa = 4.14EE310 pKa = 4.01TVLSNDD316 pKa = 3.57FEE318 pKa = 4.66NFF320 pKa = 3.09
MM1 pKa = 7.42SRR3 pKa = 11.84GKK5 pKa = 9.38SWCFTIHH12 pKa = 6.6SRR14 pKa = 11.84GEE16 pKa = 3.95GDD18 pKa = 3.41DD19 pKa = 4.79CDD21 pKa = 4.08WLLHH25 pKa = 6.3PIGTAEE31 pKa = 4.35PPSAFWDD38 pKa = 3.81DD39 pKa = 5.84DD40 pKa = 3.96PFEE43 pKa = 4.51WGCQFMIFQMEE54 pKa = 4.53RR55 pKa = 11.84CPEE58 pKa = 3.95TGKK61 pKa = 10.25LHH63 pKa = 6.41LQGALQLDD71 pKa = 3.82KK72 pKa = 10.83QQRR75 pKa = 11.84LSFMKK80 pKa = 10.32KK81 pKa = 7.85LHH83 pKa = 6.07KK84 pKa = 8.13TAHH87 pKa = 5.58WEE89 pKa = 4.07VMKK92 pKa = 10.98GNWTQSIEE100 pKa = 4.17YY101 pKa = 9.49CSKK104 pKa = 11.16SEE106 pKa = 4.15TKK108 pKa = 10.89VNGPWEE114 pKa = 4.34AGDD117 pKa = 3.8RR118 pKa = 11.84PKK120 pKa = 10.32MGQRR124 pKa = 11.84TDD126 pKa = 3.38LEE128 pKa = 4.9TIGKK132 pKa = 7.56MVKK135 pKa = 9.52EE136 pKa = 3.98NKK138 pKa = 9.25TNLEE142 pKa = 4.08LVDD145 pKa = 3.65TLGAGVSKK153 pKa = 9.79FQKK156 pKa = 9.99HH157 pKa = 4.44ISFLRR162 pKa = 11.84FTYY165 pKa = 9.9SEE167 pKa = 4.3KK168 pKa = 11.05DD169 pKa = 3.03SDD171 pKa = 3.85RR172 pKa = 11.84QATGVKK178 pKa = 10.15VIVLYY183 pKa = 11.07GPTGTGKK190 pKa = 8.99TYY192 pKa = 10.95AAVNSLTGGRR202 pKa = 11.84AYY204 pKa = 10.49HH205 pKa = 6.72ILNAPSQKK213 pKa = 9.11ATKK216 pKa = 9.64LWFDD220 pKa = 3.52GYY222 pKa = 10.92EE223 pKa = 3.98NQKK226 pKa = 10.26VLVIDD231 pKa = 4.82DD232 pKa = 4.01FDD234 pKa = 6.0GSVEE238 pKa = 3.85FRR240 pKa = 11.84YY241 pKa = 9.93LLRR244 pKa = 11.84MIDD247 pKa = 3.69VYY249 pKa = 11.21KK250 pKa = 10.75FPAEE254 pKa = 4.31VKK256 pKa = 10.45GGMVWGVWDD265 pKa = 3.82TVIITSNVHH274 pKa = 5.02PASWYY279 pKa = 8.69TGVDD283 pKa = 3.22TSPLKK288 pKa = 10.61RR289 pKa = 11.84RR290 pKa = 11.84IAEE293 pKa = 3.62IRR295 pKa = 11.84LCEE298 pKa = 3.93NQGTYY303 pKa = 11.17KK304 pKa = 9.94MISWEE309 pKa = 4.14EE310 pKa = 4.01TVLSNDD316 pKa = 3.57FEE318 pKa = 4.66NFF320 pKa = 3.09
Molecular weight: 36.57 kDa
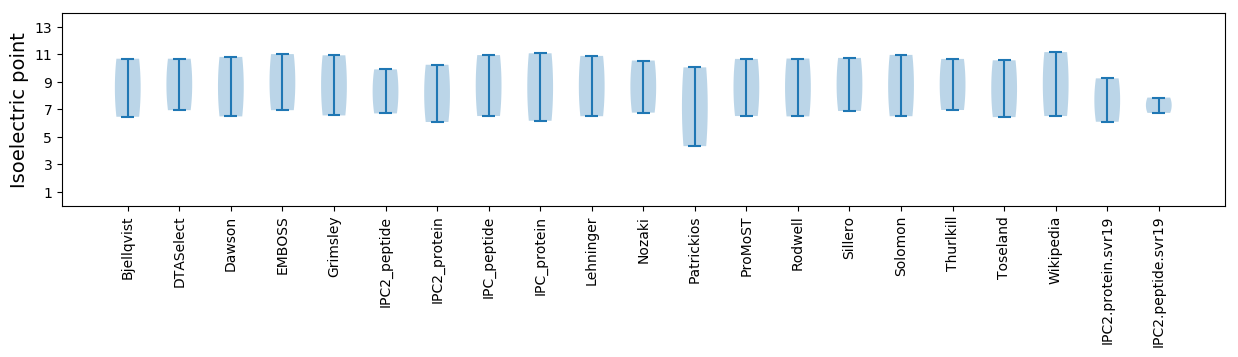
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQQ8|A0A140AQQ8_9VIRU Coat protein OS=Lake Sarah-associated circular virus-42 OX=1685771 PE=4 SV=1
MM1 pKa = 7.5MYY3 pKa = 10.22SQGAMSQRR11 pKa = 11.84RR12 pKa = 11.84TRR14 pKa = 11.84TYY16 pKa = 9.09GARR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 8.8PYY23 pKa = 10.03RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84TTSLYY31 pKa = 9.54RR32 pKa = 11.84SPYY35 pKa = 8.0TRR37 pKa = 11.84RR38 pKa = 11.84QDD40 pKa = 3.41MVPTTQGWSFNRR52 pKa = 11.84LNRR55 pKa = 11.84SSGGAQRR62 pKa = 11.84VEE64 pKa = 4.02WKK66 pKa = 10.55FSDD69 pKa = 3.57RR70 pKa = 11.84YY71 pKa = 10.44EE72 pKa = 4.83IGLTTMPLIGQFYY85 pKa = 9.85LLNGLVPGSGASQRR99 pKa = 11.84IGTMITMKK107 pKa = 10.03TVEE110 pKa = 4.6TRR112 pKa = 11.84AIWIPFSSATIGLFRR127 pKa = 11.84FFIVLDD133 pKa = 3.67HH134 pKa = 6.17QANGVTPLLSDD145 pKa = 3.33IMAATGIQFASGLRR159 pKa = 11.84NLNTRR164 pKa = 11.84QRR166 pKa = 11.84YY167 pKa = 9.02RR168 pKa = 11.84ILMDD172 pKa = 2.96KK173 pKa = 10.46TYY175 pKa = 11.07GFNNPNFTTSQPYY188 pKa = 9.94YY189 pKa = 7.4MHH191 pKa = 7.03HH192 pKa = 6.03YY193 pKa = 9.08MRR195 pKa = 11.84FRR197 pKa = 11.84LPVTTQYY204 pKa = 11.69NSGSAGSVGDD214 pKa = 3.51IVTNALYY221 pKa = 10.26FIATTTAAASATPSFQMLCRR241 pKa = 11.84LRR243 pKa = 11.84YY244 pKa = 8.18TDD246 pKa = 3.33AA247 pKa = 5.99
MM1 pKa = 7.5MYY3 pKa = 10.22SQGAMSQRR11 pKa = 11.84RR12 pKa = 11.84TRR14 pKa = 11.84TYY16 pKa = 9.09GARR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 8.8PYY23 pKa = 10.03RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84TTSLYY31 pKa = 9.54RR32 pKa = 11.84SPYY35 pKa = 8.0TRR37 pKa = 11.84RR38 pKa = 11.84QDD40 pKa = 3.41MVPTTQGWSFNRR52 pKa = 11.84LNRR55 pKa = 11.84SSGGAQRR62 pKa = 11.84VEE64 pKa = 4.02WKK66 pKa = 10.55FSDD69 pKa = 3.57RR70 pKa = 11.84YY71 pKa = 10.44EE72 pKa = 4.83IGLTTMPLIGQFYY85 pKa = 9.85LLNGLVPGSGASQRR99 pKa = 11.84IGTMITMKK107 pKa = 10.03TVEE110 pKa = 4.6TRR112 pKa = 11.84AIWIPFSSATIGLFRR127 pKa = 11.84FFIVLDD133 pKa = 3.67HH134 pKa = 6.17QANGVTPLLSDD145 pKa = 3.33IMAATGIQFASGLRR159 pKa = 11.84NLNTRR164 pKa = 11.84QRR166 pKa = 11.84YY167 pKa = 9.02RR168 pKa = 11.84ILMDD172 pKa = 2.96KK173 pKa = 10.46TYY175 pKa = 11.07GFNNPNFTTSQPYY188 pKa = 9.94YY189 pKa = 7.4MHH191 pKa = 7.03HH192 pKa = 6.03YY193 pKa = 9.08MRR195 pKa = 11.84FRR197 pKa = 11.84LPVTTQYY204 pKa = 11.69NSGSAGSVGDD214 pKa = 3.51IVTNALYY221 pKa = 10.26FIATTTAAASATPSFQMLCRR241 pKa = 11.84LRR243 pKa = 11.84YY244 pKa = 8.18TDD246 pKa = 3.33AA247 pKa = 5.99
Molecular weight: 28.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
567 |
247 |
320 |
283.5 |
32.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.82 ± 0.908 | 1.235 ± 0.514 |
4.762 ± 1.193 | 4.233 ± 1.868 |
4.938 ± 0.201 | 8.113 ± 0.26 |
1.94 ± 0.449 | 4.938 ± 0.201 |
4.938 ± 2.305 | 7.231 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.88 ± 0.606 | 3.88 ± 0.355 |
4.056 ± 0.246 | 4.586 ± 0.419 |
7.055 ± 2.149 | 7.055 ± 0.645 |
8.995 ± 1.449 | 5.115 ± 0.91 |
2.646 ± 0.886 | 4.586 ± 1.171 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
